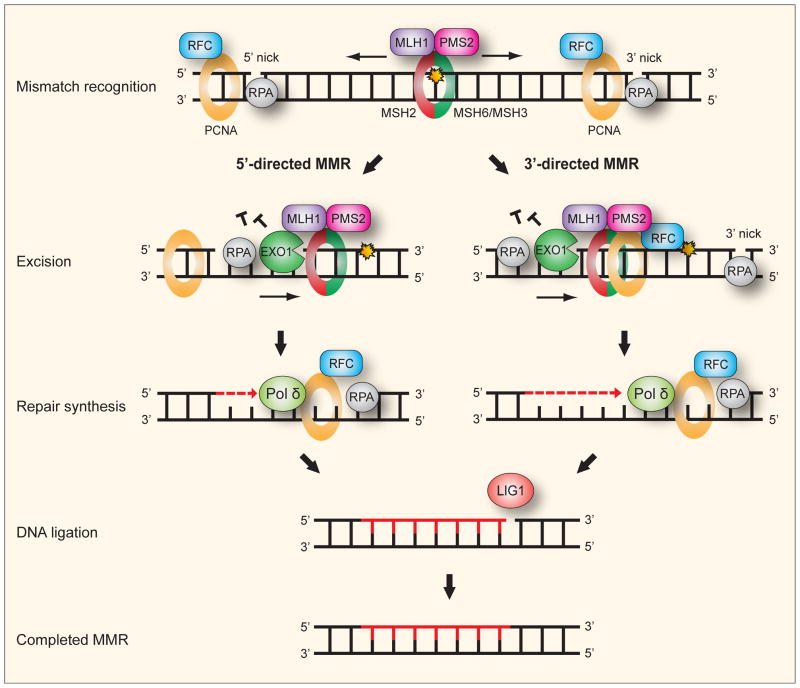
Fig. 5. Human mismatch repair.
For convenience, the mechanism of human mismatch repair (MMR) can be seen as consisting of five consecutive steps: (i) Recognition and binding of a mismatch is by either a MSH2-MSH6 or MSH2-MSH3 heterodimeric ATPase complex. MSH2-MSH6 preferentially recognizes base-base mismatches and insertion deletion loops of 1–2 nucleotides while MSH2-MSH3 has preference for larger insertion-deletion loops. The mismatch-bound MSH2-MSH6 (or MSH2-MSH3) recruits the MLH1-PMS2 complex, a molecular matchmaker with weak ATPase activity, to form a ternary complex. The PCNA clamp recruits MMR proteins to the replication fork while the clamp loader RFC loads PCNA. A strand-specific nick or gap, which may reside either 5′ or 3′ to the mismatch, is sufficient to direct repair in 5′- and 3′-directed MMR, respectively. PCNA appears essential for 3′-directed but not 5′-directed MMR. (ii) Excision is apparently by the 5′ to 3′ exonuclease EXO1 in both 3′- and 5′-directed MMR. For 5′-directed MMR the excision is straightforward by the 5′ to 3′ exonuclease activity of EXO1. For 3′-directed MMR, the endonuclease function of PMS2 is activated by presence of the 3′ nick, and stimulated by RFC, PCNA and ATP, to introduce a necessary second nick 5′ to the mismatch. Excision can then follow by EXO1. RPA binds to protect the single-stranded DNA during the excision and to facilitate the following DNA repair synthesis. (iv) Repair synthesis is accurately performed by Pol δ. (v) Ligation of the remaining nicks after synthesis is by LIG1.