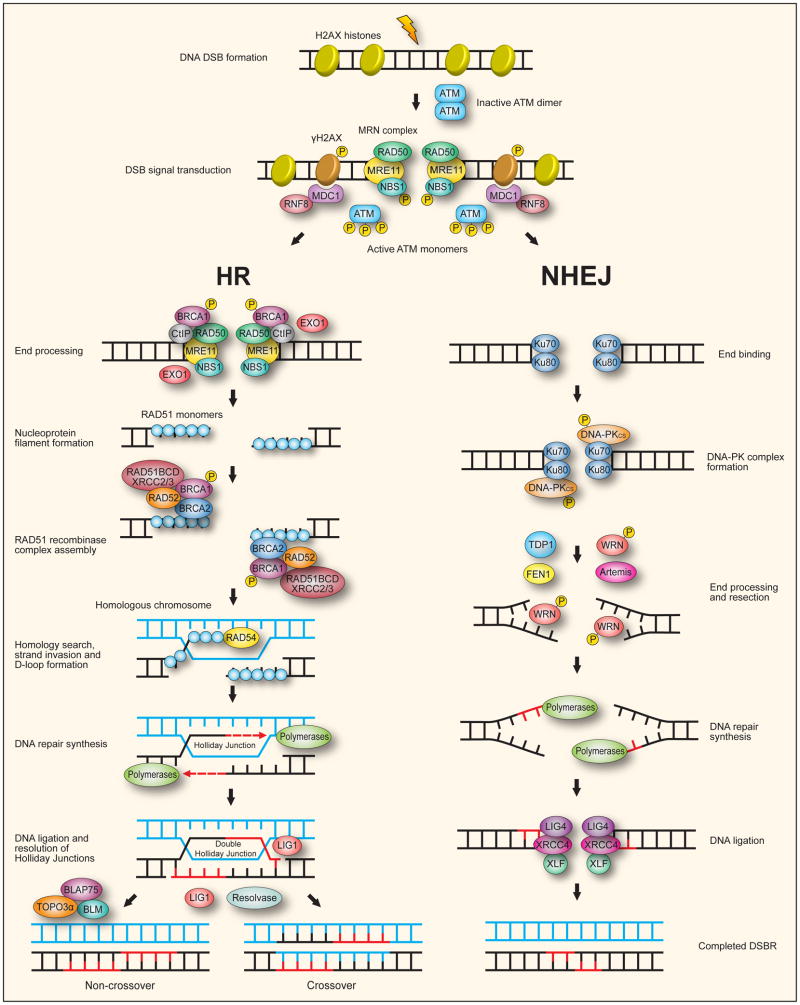
Fig. 8. The two major mammalian pathways for double-strand break repair.
Repair of a DNA double-strand break (DSB) is usually accomplished by either homologous recombination (HR) when a homologous chromosome is available in the form of a sister chromatid, or by non-homologous end-joining (NHEJ) throughout the cell cycle. Following formation of a DSB, the MRE11-RAD50-NBS1 (MRN) complex is recruited to the break where it binds to the DNA ends through MRE11. A coiled-coil region of RAD50 reaches across the break and through coordination of a central Zn2+ ion, tethers the two broken DNA ends together. The serine/threonine protein kinase ATM is usually present as an inactive dimer but is recruited to the break site by NBS1. This causes ATM to autophosphorylate (P) and monomerize, activating the kinase. The activated ATM phosphorylates NBS1 and a multitude of other proteins that participates in the DNA DSB response, including BRCA1 and phosphorylation of nearby histone H2AX to generate γH2AX. The γH2AX at repair foci acts as a signal to recruit other repair proteins in order to assemble DSBR complexes. One such protein, MDC1, binds to γH2AX which recruits RNF8to the break site where it initiates an ubiquitylation cascade of histones H2A and H2AX causing chromatin restructuring and generation of binding sites for further protein factors. The HR pathway proceeds via the end processing of damaged DNA termini with initial nucleolytic 5′ resection performed by CtIP, a function dependent on CtIP recruitment of BRCA1. Further resection by EXO1 generates single DNA strands with 3′ overhangs upon which RAD51 monomers attach to form nucleoprotein filaments. A RAD51 recombinase complex is then assembled containing the accessory proteins BRCA1, BRCA2, RAD52 and the RAD51 paralogs: RAD51B/C/D and XRCC2/3. This recombinase complex, with the aid of an additional factor, RAD54, facilitates homology search and strand invasion of the homologous chromosome to form displacement loops (D-loops). DNA repair synthesis by DNA polymerase using the homologous strand as template extends the 3′ invading strand allowing branch migration of the Holliday junction, a cruciform intermediate. The repair synthesis allows capture of the other DNA end to create a double Holliday junction intermediate. DNA ligation by LIG1 seals the remaining nicks and resolution of the double Holliday junction by structure-specific endonucleases generate either non-crossover or crossover products depending on where the junctions are cut by the resolvase. The BLM RecQ helicase can cooperate with TOPO3α and BLAP75 to resolve DHJs with generation of exclusively non-crossover products.
The NHEJ pathway first involves binding of Ku70-80 heterodimer to the two DNA termini. This then recruits DNA-PKCS to form the DNA-PK complex bringing the two DNA termini close together. Because of associated lesions not all DNA termini are readily ligatable and must be processed to generate the 5′-P and 3′-OH termini that are necessary for ligation. Autophosphorylation of DNA-PKCS can make the obstructive termini accessible to end processing enzymes such as TDP1. DNA-PKCS also mediates a regulatory phosphorylation of the WRN RecQ helicase. End processing and resection, while not fully understood, may involve the exonuclease activity of FEN1, WRN and Artemis. DNA polymerases then perform any necessary DNA repair synthesis. Finally, the DNA-PK complex, in a Ku70-80 mediated fashion, recruits the LIG4-XRCC4-XLF complex to perform the ligation of the DNA termini via the ligase activity of LIG4.