Figure 1.
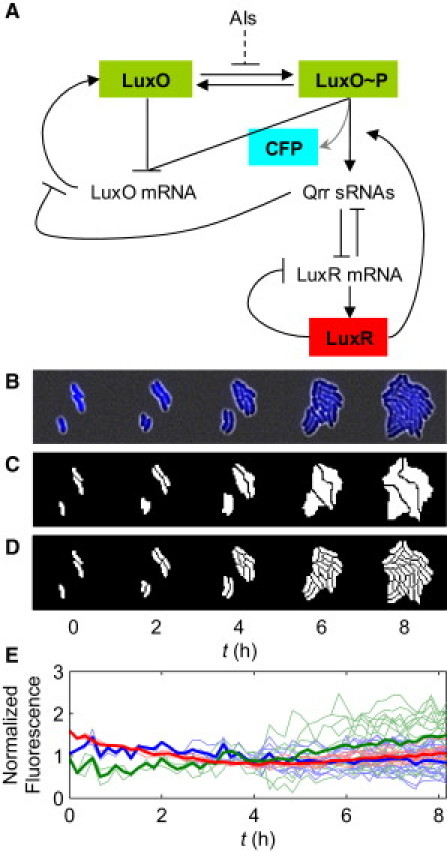
A portion of the QS circuit in V. harveyi is shown along with time-lapse images of gene expression within microcolonies. (A) A portion of the QS pathway in V. harveyi. The concentration of AIs regulates the phosphorylation of the LuxO protein. LuxO∼P activates production of the Qrr sRNAs, which repress LuxR protein production by binding and degrading its mRNA. Together with LuxO∼P, LuxR activates the production of Qrr sRNAs. LuxO represses its own transcription regardless of its phosphorylation status. Qrr sRNAs repress LuxO protein production via direct basepairing with luxO mRNA. Most lines denote regulation native to V. harveyi. For imaging purposes, LuxO is fused to YFP, and LuxR is fused to mCherry. CFP is produced together with Qrr4 (gray arrow). (B) Typical time-lapse images of V. harveyi cells. This series shows the cyan fluorescence channel (qrr4-cfp) overlaid with the white-light channel. (C) Image partitioned at the microcolony level. The three cells at the beginning of image acquisition grew into three microcolonies. (D) Image partitioned at the single-cell level. Pixels denoting cells or microcolonies are shown in white; background pixels are shown in black. (E) Typical time traces of fluorescence per unit area within a microcolony in different channels: qrr4-cfp (blue), YFP-LuxO (green), and mCherry-LuxR (red). The normalized fluorescence values are very close to unity over time. The thick curves are the microcolony averages, and the thin curves are from individual cells within the microcolony.
