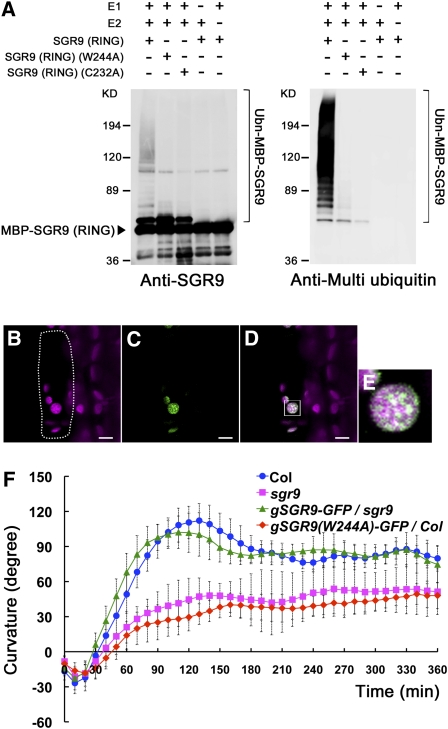
Figure 2.
Characterization of the SGR9 Protein.
(A) E3 ligase activity of the wild type and mutated SGR9 (RING). For the ubiquitination assay, MBP-SGR9 (RING), which is MBP fused to the RING finger domain of SGR9, was prepared, and amino acid substitutions W244A or C232A were introduced into the RING finger domain. An anti-SGR9 antibody was used to detect MBP-SGR9 (RING) or the mutated MBP-SGR9 (RING) (left panel), and an anti-multi ubiquitin antibody was used to detect ubiquitinated MBP-SGR9 (RING) or mutated MBP-SGR9 (RING) (right panel).
(B) to (E) Intracellular localization of mutated SGR9 protein in an endodermal cell of an inflorescence stem. Bars = 5 μm.
(B) Autofluorescence of a plastid chlorophyll. Dotted line indicates a single endodermal cell.
(C) SGR9(W244A)-GFP.
(D) Overlay of autofluorescence and GFP signal.
(E) Inset in (D). Note that mutated SGR9 protein is localized to the amyloplasts.
(F) Time course of gravitropic response of an inflorescence stem of gSGR9-GFP/sgr9 or gSGR9(W244A)-GFP/Col. Error bars represent sd. A minimum of six individuals was analyzed per sample point.