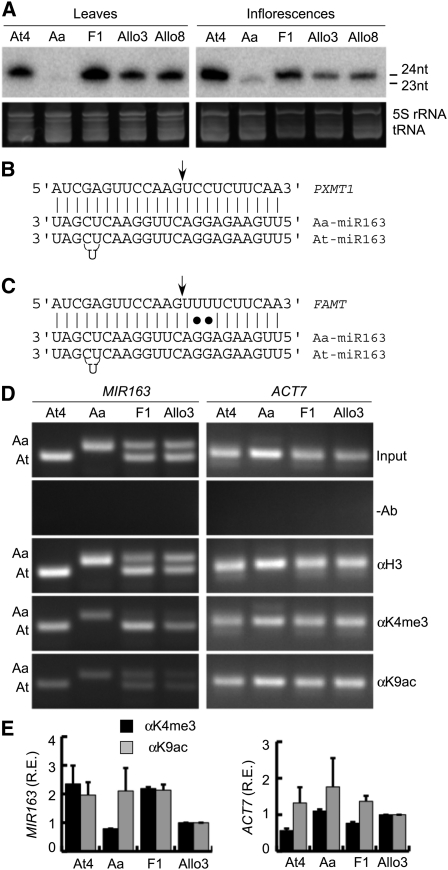
Figure 1.
Differential Accumulation of miR163 in Arabidopsis Species and Transcriptional Regulation of MIR163 by ChIP Analysis.
(A) Small RNA gel blots showing miR163 accumulation in mature leaves and inflorescences in tetraploid A. thaliana (At4), A. arenosa (Aa), and F1 and F8 (Allo3 and Allo8) generations of resynthesized allotetraploids. nt, nucleotide.
(B) Core sequence alignment of 24-nucleotide At and 23-nucleotide Aa miR163 with the target PXMT1.
(C) Core sequence alignment of 24-nucleotide At and 23-nucleotide Aa miR163 with the target FAMT.
(D) ChIP assays using antibodies against C terminus of histone H3 (H3), acetyl H3K9 (K9ac), and trimethyl H3K4 (K4me3). A negative control without antibody (-Ab) and a positive control with input DNA without IP (input) are included. Representative PCR results from ChIP and input (control) DNA of MIR163 promoter regions (266 bp in At and 286 bp in Aa) and the 5′ coding region of ACT7 (254 bp) are shown.
(E) Relative enrichment (R.E.) of MIR163 and ACT7 upstream regions for K4me3 (black) and K9ac (gray) was quantified by qPCR and normalized with the corresponding input DNA and histone H3 levels. Values are mean ± sd (from three biological replications).