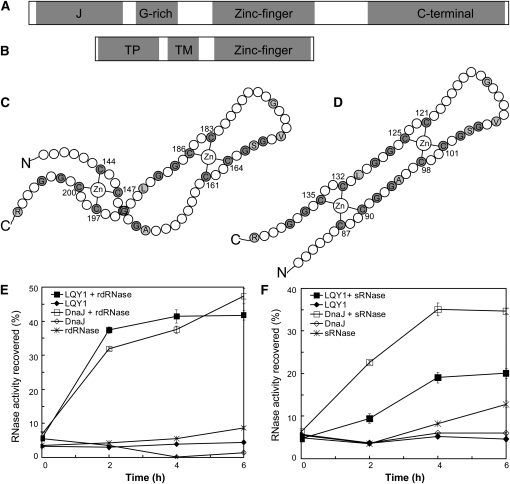
Figure 7.
The LQY1 Zn Finger Domain Has Protein Disulfide Isomerase Activity.
(A) Domains in E. coli DnaJ. The protein contains a J-domain, a Gly-rich motif, a Zn finger domain, and a C-terminal region.
(B) Domains inferred from the Arabidopsis LQY1 sequence. The protein contains a transit peptide (TP), a transmembrane region (TM), and a Zn finger domain according to in silico analysis.
(C) The topology of the E. coli DnaJ protein (Shi et al., 2005). Cys and Gly residues are shown in gray. Other residues conserved between LQY1 and DnaJ Zn finger are shown in single-letter amino acid codes.
(D) Putative topology of Arabidopsis LQY1 protein based on its homology to E. coli DnaJ.
(E) Assays for oxidative renaturation of rdRNase by recombinant LQY1 Zn finger. Restoration of activity to inactive reduced and denatured RNase A was measured for LQY1 and positive-control E. coli DnaJ. The values (mean ± se, n = 4) are normalized by the activity of native RNase A. Filled squares, LQY1 and rdRNase; filled diamonds, LQY1, no rdRNase; open squares, DnaJ and rdRNase; open diamonds, DnaJ, no rdRNase; asterisks, rdRNase, no LQY1 or DnaJ.
(F) Reductive renaturation of sRNase by recombinant LQY1 Zn finger. The values (mean ± se, n = 4) are normalized as in (E). Filled squares, LQY1 and sRNase; filled diamonds, LQY1, no sRNase; open squares, DnaJ and sRNase; open diamonds, DnaJ, no sRNase; asterisks, sRNase, no LQY1 or DnaJ.