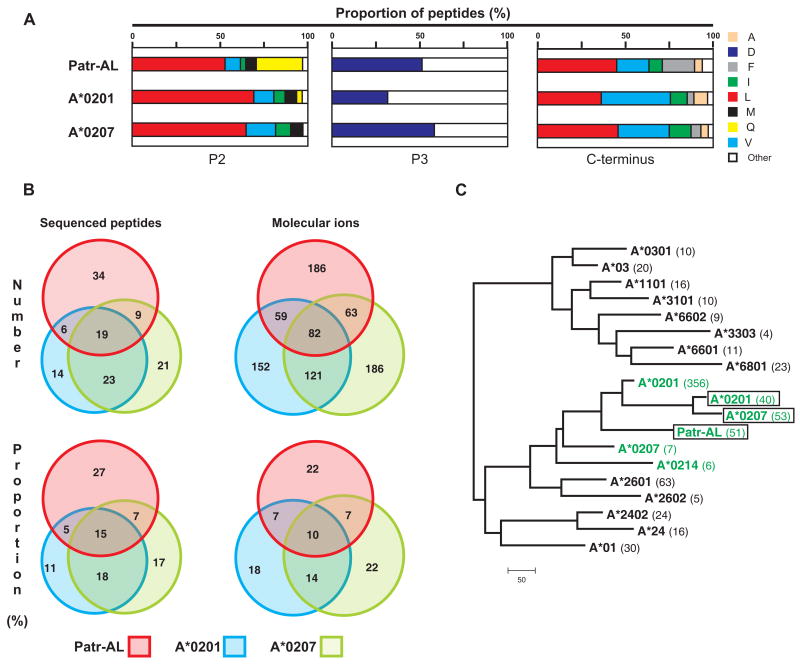
Figure 3. Extensive overlap between the repertoires of peptides bound by Patr-AL and HLA-A*02.
(A) Distribution of amino acid residues at the three anchor positions of the bound peptide: position 2 (P2), position 3 (P3), and the C-terminus. Proportions were calculated from the amino-acid sequences of 126 binding peptides for Patr-AL, HLA-A*0201 and HLA-A*0207. (B) Overlap of the peptide repertoires bound by Patr-AL, HLA-A*0201 and HLA-A*0207. The peptides binding to each MHC class I are defined by the differently colored circles: Patr-AL, red; A*0201, blue; and A*0207, green. The four overlapping regions between the circles defines the peptides bound by all three MHC class I and by the three combinations of two of them. On the left under ‘Sequenced peptides’ is shown the analysis for the peptides for which the amino-acid sequences are known. On the right under ‘Molecular ions’ is shown a second, independent study in which peptides were defined by the weight of their molecular ions. (C) Neighbor-joining phylogenetic tree to compare the peptide-binding specificities of Patr-AL, A*0201, and A*0207 as defined in our analysis (green and boxed), with those previously defined for A*0201, A*0207, A*0214 (green) and sixteen other HLA-A allotypes obtained from the SYFPEITHI database (39). The number of peptides in each dataset is shown in parentheses following the allotype name.