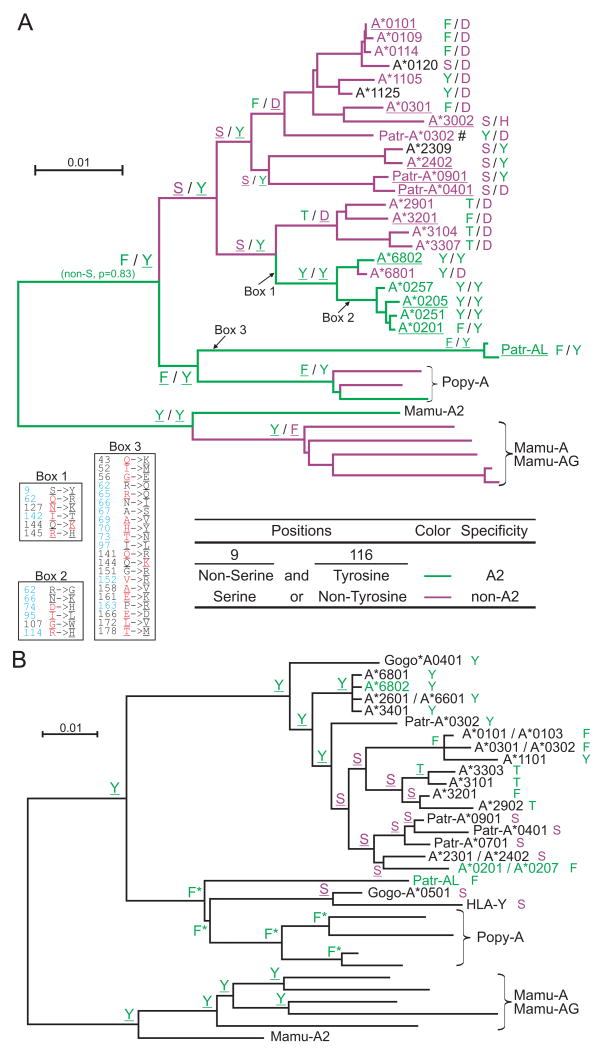
Figure 7. Evolution of the A2 peptide-binding specificity at the HLA-A gene.
Combination of residues at positions 9 and 116 of the MHC-A and -AL peptide binding domain correlates with the presence or absence of the A2 peptide specificity (summarized in Fig. 6). Panel A shows the results of ancestral sequence reconstruction for both positions on the phylogenetic tree shown in Fig. 2. To complement this analysis, and extend the set of sequences that could be included in the analysis, we generated a phylogenetic tree on a smaller gene segment beginning 300bp upstream of the ATG start codon and ending in exon 2 (supplemental Fig. S6) and used this tree for ancestral reconstruction of position 9 (panel B). (A) The identities at positions 9 and 116 from the ancestral sequence reconstruction are shown for thirteen nodes. The branches of the tree and the names of the sequences are colored according to peptide specificity (green for A02 and purple for non-A02): for the branches, peptide specificity is based on the residues predicted at positions 9 and 116. For contemporary allotypes, the specificities were obtained from Sidney et al. (68) (underlining indicates ‘observed’ specificities). For each sequence the residues at positions 9 and 116 are shown to the right. The amino acid changes that occurred along three branches are given in boxes: positions colored in blue represent peptide-binding sites (89), residues colored in red along the HLA-A2 branches represent residues found in Patr-AL, while residues colored in red along the Patr-AL branch represent HLA-A*0201 residues. Underlined residues have p>0.95. #, specificity is based on Patr-A*0301. (B) Identity of position 9 from the ancestral sequence reconstruction is shown for all nodes. For each sequence the residue at position 9 is shown to the right. Serine is colored purple, all others are green. Names of MHC class I with A02 peptide specificity are green. Underlined residues have p>0.95. *, non-serine residue (p=1.0).