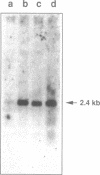
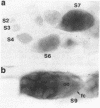
Abstract
Proteins of the D-E-A-D family of putative ATP-dependent RNA helicases have been implicated in translation initiation and RNA splicing in a variety of organisms from E. coli to man. The Drosophila vasa protein, a member of this family, is required in the female germ line for fertility and for specification of germ line and posterior positional information in progeny embryos. We report the isolation of another D-E-A-D gene from Drosophila, which, like vasa, is expressed in germ line tissue. The predicted amino acid sequence of this new gene, Dbp73D, contains all of the highly conserved helicase motifs, but is otherwise the farthest-diverged member of the family so far identified.
Full text
PDF
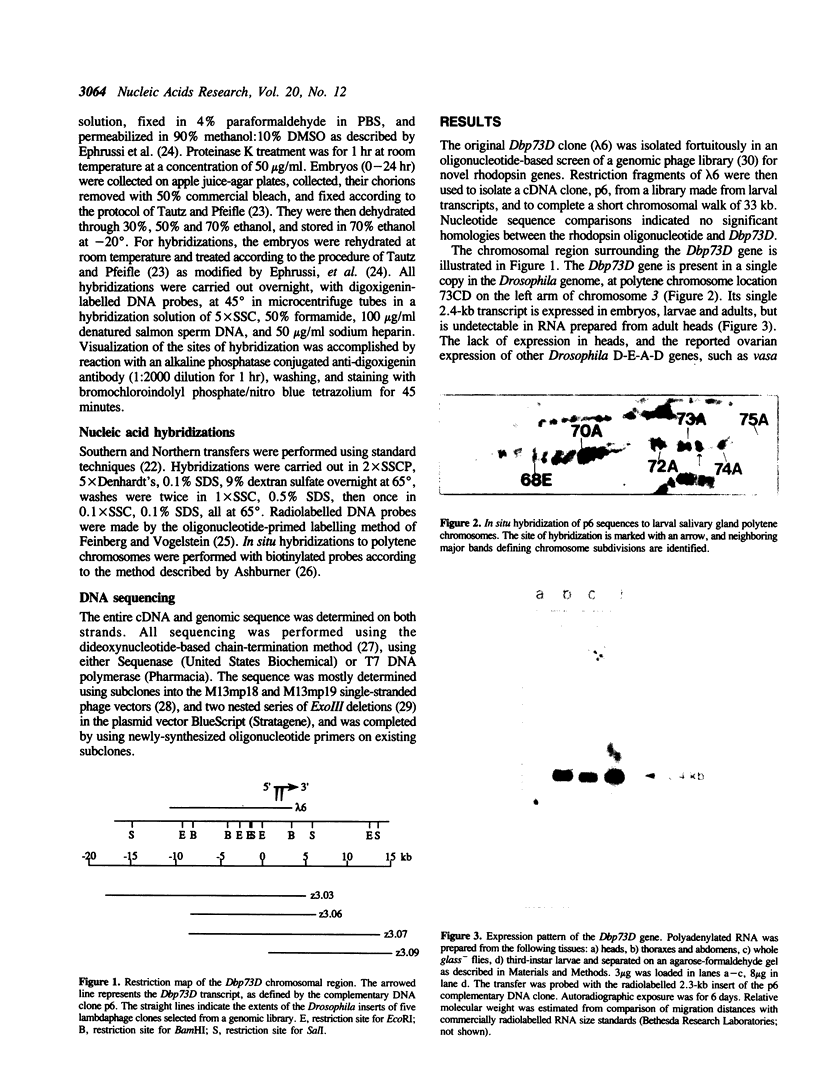
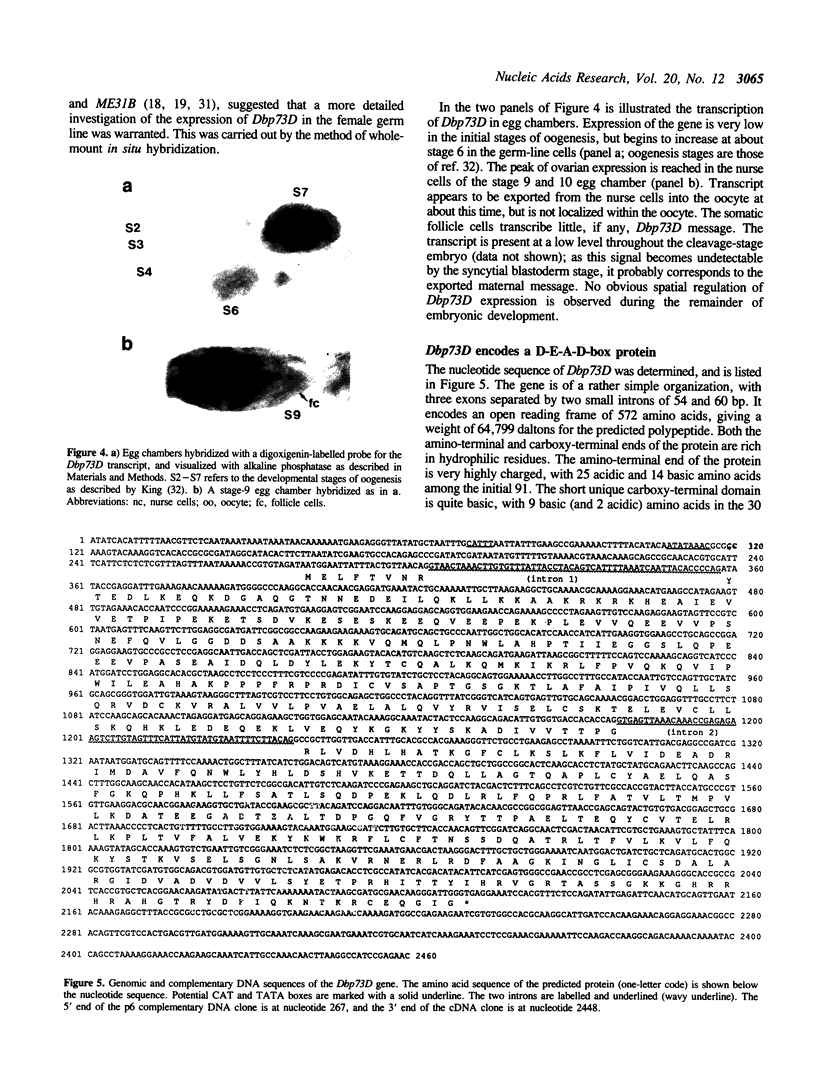


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballinger D. G., Benzer S. Targeted gene mutations in Drosophila. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9402–9406. doi: 10.1073/pnas.86.23.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr P. J. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991 Jul 12;66(1):1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Belote J. M., Hoffmann F. M., McKeown M., Chorsky R. L., Baker B. S. Cytogenetic analysis of chromosome region 73AD of Drosophila melanogaster. Genetics. 1990 Aug;125(4):783–793. doi: 10.1093/genetics/125.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Couto J. R., Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990 Mar 9;60(5):705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- Chang T. H., Arenas J., Abelson J. Identification of five putative yeast RNA helicase genes. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1571–1575. doi: 10.1073/pnas.87.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. H., Lin R. J. The yeast PRP2 protein, a putative RNA-dependent ATPase, shares extensive sequence homology with two other pre-mRNA splicing factors. Nucleic Acids Res. 1990 Nov 11;18(21):6447–6447. doi: 10.1093/nar/18.21.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Arenas J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991 Feb 7;349(6309):487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hay B., Ackerman L., Barbel S., Jan L. Y., Jan Y. N. Identification of a component of Drosophila polar granules. Development. 1988 Aug;103(4):625–640. doi: 10.1242/dev.103.4.625. [DOI] [PubMed] [Google Scholar]
- Hay B., Jan L. Y., Jan Y. N. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988 Nov 18;55(4):577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Jamieson D. J., Rahe B., Pringle J., Beggs J. D. A suppressor of a yeast splicing mutation (prp8-1) encodes a putative ATP-dependent RNA helicase. Nature. 1991 Feb 21;349(6311):715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- Kaiser K., Goodwin S. F. "Site-selected" transposon mutagenesis of Drosophila. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1686–1690. doi: 10.1073/pnas.87.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J., Smith J. L., Macdonald P. M. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991 Jul 12;66(1):23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- Lasko P. F., Ashburner M. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 1990 Jun;4(6):905–921. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- Lasko P. F., Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988 Oct 13;335(6191):611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lawson T. G., Lee K. A., Maimone M. M., Abramson R. D., Dever T. E., Merrick W. C., Thach R. E. Dissociation of double-stranded polynucleotide helical structures by eukaryotic initiation factors, as revealed by a novel assay. Biochemistry. 1989 May 30;28(11):4729–4734. doi: 10.1021/bi00437a033. [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas A., Montine K. S., Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5' cap. Nature. 1990 Jun 7;345(6275):544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Leroy P., Alzari P., Sassoon D., Wolgemuth D., Fellous M. The protein encoded by a murine male germ cell-specific transcript is a putative ATP-dependent RNA helicase. Cell. 1989 May 19;57(4):549–559. doi: 10.1016/0092-8674(89)90125-6. [DOI] [PubMed] [Google Scholar]
- Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., Nishi K., Schnier J., Slonimski P. P. Birth of the D-E-A-D box. Nature. 1989 Jan 12;337(6203):121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Ray B. K., Lawson T. G., Kramer J. C., Cladaras M. H., Grifo J. A., Abramson R. D., Merrick W. C., Thach R. E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985 Jun 25;260(12):7651–7658. [PubMed] [Google Scholar]
- Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990 Mar;10(3):1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. Pre-mRNA splicing in yeast. Trends Genet. 1991 Mar;7(3):79–85. doi: 10.1016/0168-9525(91)90276-V. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990 Mar 2;247(4946):1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. Cap-binding proteins of eukaryotic messenger RNA: functions in initiation and control of translation. Prog Nucleic Acid Res Mol Biol. 1988;35:173–207. doi: 10.1016/s0079-6603(08)60614-5. [DOI] [PubMed] [Google Scholar]
- Strauss E. J., Guthrie C. A cold-sensitive mRNA splicing mutant is a member of the RNA helicase gene family. Genes Dev. 1991 Apr;5(4):629–641. doi: 10.1101/gad.5.4.629. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Simon M., Boulet A., Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989 Jan 5;337(6202):84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- Tautz D., Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989 Aug;98(2):81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. RNA splicing. Alive with DEAD proteins. Nature. 1991 Feb 7;349(6309):463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Valoir T., Tucker M. A., Belikoff E. J., Camp L. A., Bolduc C., Beckingham K. A second maternally expressed Drosophila gene encodes a putative RNA helicase of the "DEAD box" family. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2113–2117. doi: 10.1073/pnas.88.6.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]