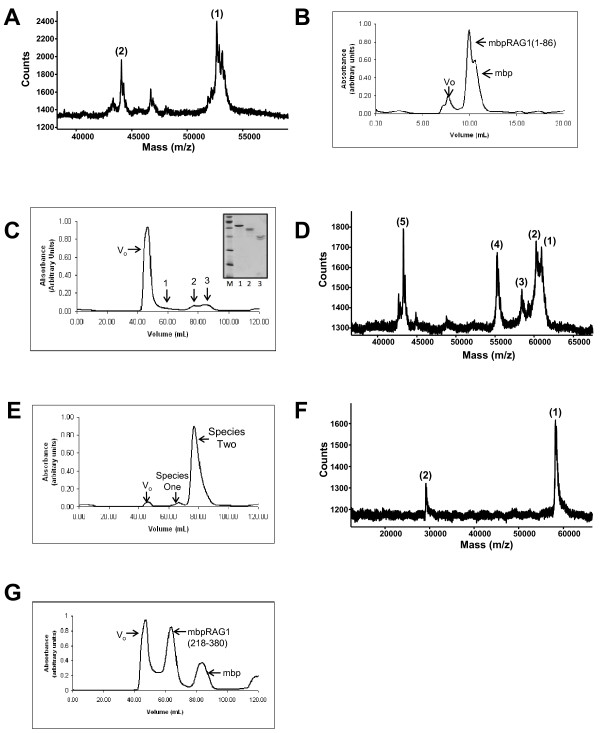
Figure 2.
Identification of domains in non-core RAG1. (A) MALDI-TOF spectrum of proteolytic products generated during purification of MBP-RAG1(1-380). Two peaks were observed with molecular weight (MWt) values: (1) 52,756.4 Da, consistent with cleavage C-terminal to residue 86, and (2) 44,061.9 Da, consistent with cleavage just C-terminal to the MBP portion of the fusion construct. The predicted mass of MBP-RAG1(1-86) is 52,815 Da. (B) SEC of MBP-RAG1(1-86). (C) SEC of MBP-RAG1(87-380). Vo denotes the void volume and the position at which aggregated protein elutes. Inset: SEC peaks 1-3 analyzed by SDS-PAGE. (All lanes are from the same gel.) Samples in lanes 1-3 correspond to peaks assigned in the SEC elution profile. (D) MALDI-TOF spectrum of peak 2 from the SEC of MBP-RAG1(87-380). Five major peaks were observed with MWt values: (1) 60,902.8, (2) 60,255.1, (3) 58,410.8, (4) 55,239.7, and (5) 43,273 Da. These values are consistent with cleavage C-terminal to residues 238, 232, 217, 190, and the MBP portion of the fusion construct, respectively. Predicted masses for the corresponding MBP-RAG1 polypeptides are 60,897 Da (MBP-RAG1(87-238)), 60,199 Da (MBP-RAG1(87-232)), 58,400 Da (MBP-RAG1(87-217)), and 55,240 Da (MBP-RAG1(87-190)). (E) SEC of MBP-CND following subcloning and purification. (F) MALDI-TOF spectrum of MBP-CND. The peaks yield MWt values consistent with the +1 (1) and +2 (2) species of the protein. (G) SEC of MBP-bZDD following subcloning and purification. All SEC runs were performed using Superdex 200 columns with either 20 ml (panel B) or 200 ml (panels C, E, and G) column volumes.