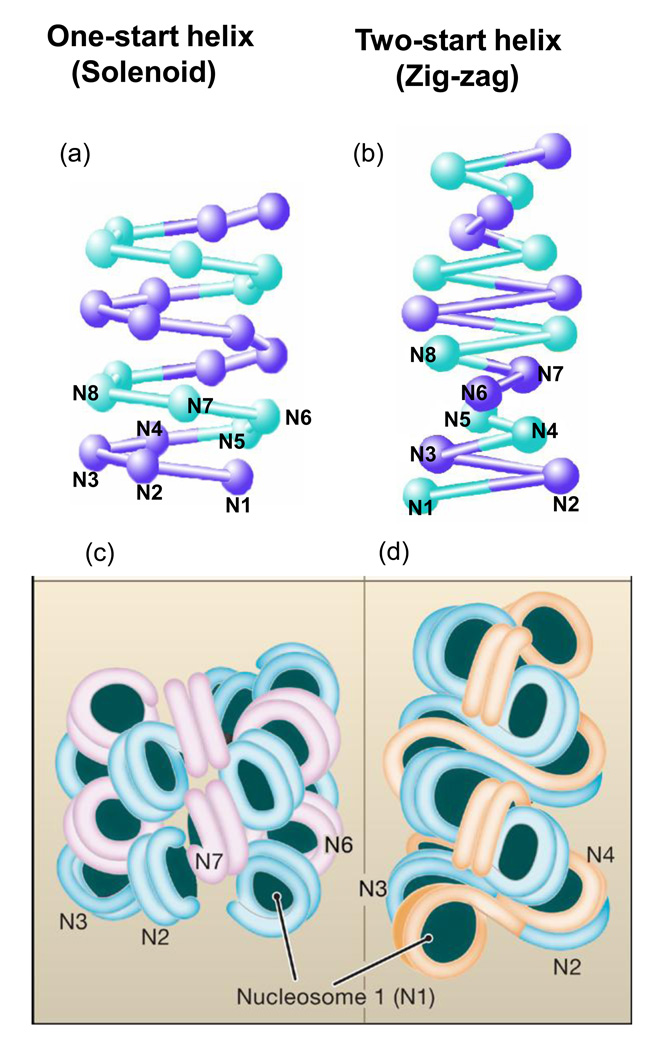
Figure 1. Models of the 30 nm Chromatin Fiber.
Two well-known structural models for 30-nm chromatin fibers are proposed: one-start helix (solenoid) (a and c) and two-start helix (zig-zag) (b and d). At the top, a schematic representation is shown for the two different topologies of 30-nm chromatin fibers (a and b). The alternative nucleosomes are numbered from N1 to N8. In the solenoid model proposed by Rhodes and colleagues, the 30-nm chromatin fiber is an interdigitated one-start helix in which a nucleosome in the fiber interacts with its fifth and sixth neighbor nucleosomes [19]. Alternative helical gyres are colored blue and magenta (c). In the zigzag model suggested by Richmond and colleagues, the chromatin fiber is a two-start helix in which nucleosomes are arranged in a zig-zag manner such that a nucleoosme in the fiber binds to the second neighbor nucleosome[18,20]. Alternative nucleosomes pairs are colored blue and orange. Image courtesy of D. Rhodes.