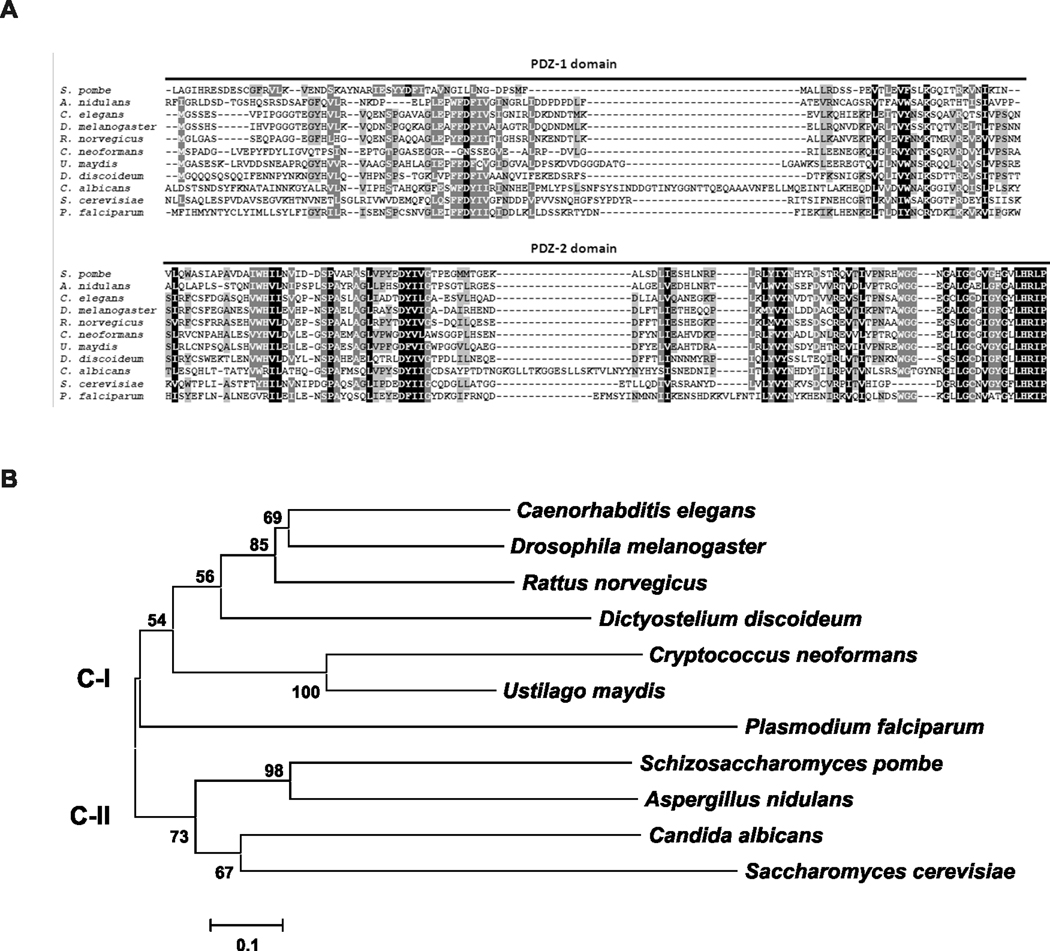
Figure 2. Phylogenetic analysis of the C. neoformans GRASP ortholog.
A. Alignment of the GRASP PDZ domains (PDZ-1 and PDZ-2) from R. norvegicus (AAB81355.2), C. elegans (NP_501354), D. melanogaster (AAF49092), S. cerevisiae (NP_010805), D. discoideum (EAL60823), S. pombe (NP_593015.1), P. falciparum (AAN35366), A. nidulans (Broad Institute accession number ANID_11248), U. maydis (Broad Institute accession number UM01076), C. albicans (Broad Institute accession number CAWG_01021) and C. neoformans (Broad Institute accession number CNAG_03291) using ClustalX2. B. Phylogenetic analysis applying the Neighbor-Joining method including GRASP sequences from distinct eukaryotic organisms listed above. The phylogeny tree splits into two major clades. C-I and C-II represents clades I and II, respectively. The bar marker indicates the genetic distance, which is proportional to the number of amino acid substitutions.