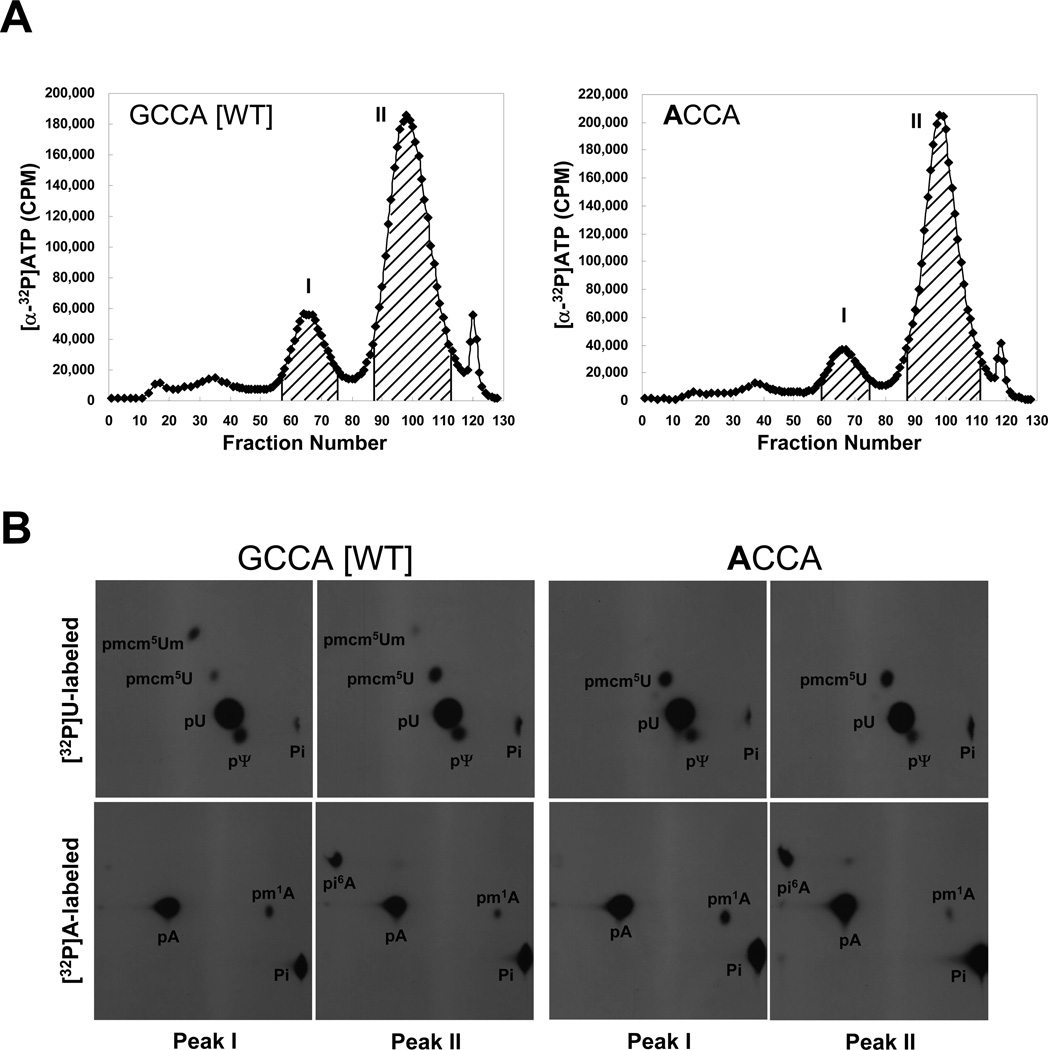
Fig. 2.
Elution profiles of 32P-labeled wild type and mutant tRNA[Ser]Sec isoforms and modified base and nucleoside analyses. In (A), [α-32P]UTP- or [α-32P]ATP-labeled wild type (GCCA [WT]) and the discriminator base (ACCA) tRNA[Ser]Sec transcripts, were prepared, microinjected into Xenopus oocytes, incubated overnight, extracted and chromatographed on a RPC-5 column as described in Materials and Methods. The graphs show the elution profiles of [α-32P]ATP labeled tRNAs (and the [α-32P]UTP elution profiles are shown in Supplementary Fig. 1). X and Y axis represent fraction number and CPM, respectively. Peaks I and II were pooled as shown by the hatched areas in the figures, collected, digested with nuclease, and in (B), the digests of the two peaks of [α-32P]UTP- or [α-32P]ATP-labeled were resolved by two-dimensional TLC as described in Materials and Methods and the modified bases and Um34 detected in the digests by autoradiography as described (see text and [4]). The films were exposed for 12–16 h.