Abstract
Genetic testing for heritable hearing loss involves a mix of patented and unpatented genes, mutations and testing methods. More than half of all hearing loss is linked to inherited mutations, and five genes are most commonly tested in the United States. There are no patents on three of these genes, but Athena Diagnostics holds exclusive licenses to test for a common mutation in the GJB2 gene associated with about 50% of all cases, as well as mutations in the MTRNR1 gene. This fragmented intellectual property landscape made hearing loss a useful case study for assessing whether patent rights in genetic testing can proliferate or overlap, and whether it is possible to gather the rights necessary to perform testing. Testing for hearing loss is widely available, primarily from academic medical centers. Based on literature reviews and interviews with researchers, research on the genetics of hearing loss has generally not been impeded by patents. There is no consistent evidence of a premium in testing prices attributable to patent status. Athena Diagnostics has, however, used its intellectual property to discourage other providers from offering some tests. There is no definitive answer about the suitability of current patenting and licensing of commonly tested genes because of continuing legal uncertainty about the extent of enforcement of patent rights. Clinicians have also expressed concerns that multiplex tests will be difficult to develop because of overlapping intellectual property and conflict with Athena’s sole provider business model.
Keywords: Patents, Intellectual Property, Hearing Loss, Deafness, Microarray Analysis, Athena Diagnostics, genetic testing
INTRODUCTION
Inherited DNA mutations account for over half of all hearing loss cases. Genetic hearing loss can be classified as “syndromic” or “nonsyndromic,” depending on whether there are associated clinical features (syndromic) or not (nonsyndromic). Mutations in a multitude of individual genes have been implicated in genetic hearing loss. In some cases, a single mutated gene is associated with hearing loss (dominant) and in others, symptoms occur when both parental genes an individual inherits are mutated (recessive) or a mutation occurs on the X chromosome (X-linked). Mutations in a few genes are the most common: GJB2/Connexin 26, GJB6/Connexin 30, SLC26A4/PDS, MTRNR1, and MTTS1. Those mutations are most commonly tested in the US.
Genetic testing for hearing loss can be controversial. Deafness and acquired hearing loss are disabilities, and whether or not to classify them as medical conditions is contested. Beliefs, lived experiences, and attitudes of individuals, both in the hearing and the Deaf Community differ widely. Whether genetic testing is useful or valuable is not a point of consensus. The complexities of when, whether, and how to classify deafness or hearing loss as a medical condition are beyond the scope of this case study. This case study is about testing for inherited mutations that can cause loss of hearing, but with no particular view about whether such testing is valuable or whether it is a medical service.
The diverse perspectives on whether hearing loss is a disease or a disability influence consumer utilization of tests.1,2 This complicates the notion of “access,” because consumer values and preferences affect utilization. For those who deliberately choose not to use tests, lack of utilization does not indicate lack of access but rather expression of a choice. While this is true in general for all genetic testing, the fact that many in the Deaf Community contest the understanding of deafness as a disability is particularly relevant to this particular case study. Statistics on utilization are always only an indirect measure of access, but for hearing loss utilization rates are particularly suspect. Access is about how many people who want information and could benefit from it can get it; how hearing loss and deafness are regarded directly affects how many people actually want to know the cause, and consequently how many people want testing. Hereafter our analysis will proceed on assumption that we are addressing the use of genetic testing among those who want it and can benefit from it, while recognizing that some would not seek testing even if it were freely available at no cost, and access were not an issue.
Clinical guidelines from the American College of Medical Genetics (ACMG) recommend incorporating genetic testing into the diagnosis of congenital hearing loss.3 The benefits of genetic testing in diagnostic evaluation of hearing loss include:
Reducing additional time-consuming and expensive testing;
Facilitating early interventions such as hearing aids, cochlear implants, or sign language that significantly improve language ability;
Understanding disease progression;
Monitoring associated clinical manifestations and complications, particularly for syndromic hearing loss; and
Providing accurate information on the chance of recurrence that some may choose to use in making decisions about having children (and others may not).
PATENT ISSUES CONCERNING A MULTI-GENE, MULTI-MUTATION CONDITION
Hearing loss provides an opportunity to investigate how the patenting of different genes, mutations, and methods by multiple parties can affect access to genetic testing. Patents on multiple DNA sequences (both normal genes and mutations) owned by many parties have raised concerns about “patent thickets” or “anticommons.” An anticommons can occur when it becomes difficult to offer a service because the intellectual property is dispersed, making it difficult to accumulate all the permissions needed to offer genetic tests for the mutations that might cause hearing loss. This problem was characterized most famously in May 1998 by Heller and Eisenberg.4
The related notion of a patent thicket is that there is so much intellectual property that needs to be accumulated that it becomes difficult to cut through it all. This is a problem of density and profusion. The two concepts are distinct, but travel together in the real world, in areas where many patents have been granted to many players.
Another concern about patents is “blocking,” where a single patent owner with claims pertaining to common variants (or to a key method) can block others from doing genetic testing. Blocking can happen from just one or a few patents on key sequences, key methods, or other inventions, if they are difficult or impossible to invent around. This is a concern for hearing loss genes because patents on one or a few common variants might enable those who hold the relevant patents to prevent others from testing for other hearing loss genes.
One concept in intellectual property that requires aggregation of many patent rights is the incentive for hold-out. This was not highlighted in our case study, but is a possibility in the future, depending on how the tests evolve. The fact that different mutations have different frequencies (and therefore explain different fractions of cases) means that the potential commercial value of a mutation patent varies. Patents covering common variants should, therefore, generally be more valuable for clinical testing than rare ones. This makes patent pooling more complicated, because many pools simply count patents rather than try to weigh their value, and this may not work for genetic testing even if all the other issues about setting up patent pools were to get resolved. The hold-out incentive appears when a pool has started to form, but a key patent lies outside the pool, and the patent-holder perceives he or she has bargaining advantage and gets a disproportionate benefit (a “hold-out premium”) for joining the pool compared to others already in. This is not distinctive to gene patents, but it could surface as a problem if patent pools begin to emerge.
The blocking effect is related to the somewhat different phenomenon of a “penumbra” effect. We characterize a penumbra as activities by a patent holder that are not strictly speaking infringing activities but that in practice result from controlling one or a few patents. This phenomenon appears in this case study because having rights to some common variants can in effect force those who want genetic testing to go to a particular single provider, even though no one can know in advance whether the mutations for which that provider has exclusive rights are actually the ones that cause symptoms in that individual.
One important purpose of seeking genetic testing for hearing loss is to identify the precise molecular cause of the symptoms. So if one testing service retains exclusive rights to test for a common variant, then everyone will of course need to test for that variant, and therefore will send samples to that service, even though the patient may actually have some other mutation—whether unpatented, discovered by someone else and patented, or that no one has ever before discovered. By having rights to one common variant, therefore, a service can force all who seek genetic testing for an entire clinical syndrome to come to it, even if its intellectual property covers only a fraction of all possible mutations. The owner of the key patent thereby controls not only his or her own intellectual property, but collateral space. This enables the holder to accumulate knowledge and expand its intellectual property. All those with hearing loss and seeking genetic testing will come to this service, and new mutations will thus be found by it, leading to more patents for mutations for that condition. By this mechanism, a monopoly on the original discovery is leveraged to future discoveries and future patents on new mutations that no one has discovered before, in the clinical penumbra of the originally patented test.
The penumbra effect in effect expands the intellectual property controlled by the initial patent holder, but it can also create some perverse incentives for subsequent inventions falling in the penumbra of the original patent. Those discovering a hearing loss mutation may think about simply leaving the discovery in the public domain. This might even be socially optimal by making the discovery available for both scientific progress and also making it easy for any testing service to incorporate the new discovery into ongoing testing. But if one service is controlling the testing because it has patent rights to common variants, then leaving the discovery unpatented merely fuels that service’s advantage. The institution making the new discovery will thus face several choices: (1) patent and license to the dominant provider, getting a piece of the action (and thus increasing costs in general, both transaction costs of getting and licensing the patent, but also the pass-through costs to the provider and even higher pass-through costs to end-users—this is the option taken by many institutions in this case study); (2) patent and nonexclusively license; (3) don’t patent and forego royalties (true for several in this case study); or (4) patent and license to an entirely different provider, setting up a mutual-blocking situation among service providers. To our knowledge mutual blocking has not occurred in this case, but it does appear to be developing for Long-QT syndrome in a separate case study. All of these options are socially suboptimal by one criterion or another (fairness, efficiency, or both). The penumbra effect is one of the reasons that diagnostic licensing will be a difficult policy problem to solve.
Finally, when a clinical condition requires testing for mutations or uses methods covered by many patents, this can increase costs due to royalty stacking (because payments to many patent owners are required). This is a common problem in technology licensing, and not distinctive to diagnostics. The solutions include having a cap on total royalties, clauses in licenses that permit royalty reduction if further licenses become necessary to practice an invention, patent pools, and renegotiation rules. These solutions are all dependent on licensing terms. Because licensing is largely opaque—those out-licensing in-licensing technologies have no obligation to share terms of licensing with us—we do not know the extent to which these issues have been addressed in patent licenses that affect genetic testing for hearing loss.
In this case study, we assess the patent status of hearing loss genes and go as far as we can in judging whether or not they pose the potential for a patent thicket, or anticommons, and also the possibility of blocking patents and the penumbra effect. To our knowledge, royalty stacking was not identified as a major problem, although some have wondered about it in interviews. Our main findings are:
Most hearing loss genes identified to date are not patented. It does not follow that testing for mutations in these genes is freely available, because of the penumbra effect.
Testing for Connexin 26 gene mutations, which account for up to half of all non-syndromic recessive hearing loss cases, is patented.
Of the five most commonly tested hearing loss genes, three (GJB6, SLC26A4, and MTTS1) are not patented. Clinical testing is offered for each of these genes by several providers listed on the GeneTests.org website.
Testing for mutations in genes involved in less common forms of hearing loss is predominantly offered on a research basis, if it is available at all. Laboratories doing genetic testing for research purposes are generally not CLIA-certified.
The Institut Pasteur holds two patents (US 5,998,147 and 6,485,908) for the GJB2/ Connexin 26 gene and for detecting its most common deletion mutation 35delG.
GJB2 patents have been exclusively licensed, apparently with territory of use restrictions, to the for-profit company Athena Diagnostics for testing in the United States, Canada, and Japan. (The documents that specify terms of licensing, including territorial restrictions, are not public, so we can only infer such terms.)
Cedars-Sinai Medical Center holds a patent (US 5,506,101) that covers MTRNR1 mutation testing, specifically testing for the most common A1555G mutation. This patent is also exclusively licensed to Athena Diagnostics.
LESSONS LEARNED
Research
Research on both rare and common forms of hearing loss appears to have progressed independently of patenting status. There is no evidence that patents have had any positive or negative impact on hearing loss genetics research.
Research on microarray and chip-based diagnostics for hearing loss is being performed by multiple groups. These diagnostics include patented genes and mutations and are currently offered on a research-only basis in the U.S.
Concerns about increased patent enforcement have been raised by some researchers, who worry about both research and clinical access.
Development and Commercialization
We found no evidence that patents accelerated or inhibited hearing loss test development.
Diagnostic tests for both patented (GJB2, MTRNR1) and unpatented genes (SLC26A4, GBJ6, and MTTS1) have been developed and are offered as a clinical service by several providers. Demand for testing and the extent of research on hearing loss appear to be the primary factors that determine whether diagnostic testing for a particular hearing loss gene is offered as a clinical service at that institution.
Several providers offer testing panels that include both patented and unpatented tests, e.g., GJB2/Cx26 and GJB6/Cx 30 and MTRNR1 and MTTS1.
Testing for GJB2 mutations, which is licensed exclusively to Athena Diagnostics in the U.S., was offered as early as 1998. At least 19 providers offered the test in the U.S in January 2009, a majority of which are academic medical centers. However there have been intermittent enforcement efforts by Athena Diagnostics and some laboratories have stopped testing. In August 2008 one provider (Boston University’s Center for Human Genetics) stopped offering Connexin 26 and MTRNR1 testing following Athena’s enforcement actions. The recent discontinuation of ASRs offered by Third Wave Technologies to detect the 35delG mutation has increased concern about inability to circumvent patents covering 35delG mutation detection controlled by Athena. (Third Wave Technologies Inc was acquired by HoloLogics Inc. in June 2008 and has discontinued marketing several ASRs for genetic testing, including Connexin 26 mutation testing, for business reasons.5) This may change the number of providers offering GJB2 testing. Laboratories previously using a two-tiered approach for GJB2 testing, first detecting the 35delG mutation with the ThirdWave Invader™ assay, followed by full sequencing, especially if the sample is negative for 35delG mutations, may now be prevented from reporting out 35delG mutations. This may limit providers from performing clinically meaningful testing since 35delG is the most common GJB2 mutation and some providers may stop offering the test altogether, especially if Athena steps up enforcement activity.
The price of genetic tests for hearing loss does not appear to correlate with patent status alone. The most expensive test is for Pendred Syndrome, and involves full sequence analysis of SLC26A4. There are no patents associated with the SLC26A4 gene and average test price is ~$1,700. In contrast, testing for GJB2, which is patented, has a list price ranging from $336 to $818. However the price per amplicon for full sequence analysis of GJB2 ($140.8– $430/per amplicon) appears to be higher than SLC26A4 sequencing prices, which range from $55.00–$125.25/per amplicon. This price differential cannot be attributed to patents or licensing, however, because most providers of GJB2 testing probably do not have sublicenses from Athena. (Athena states it has not issued sublicenses.) Factors such as how labor and fixed costs are distributed in test pricing may contribute to this price difference.
The cost for GJB2 full sequence analysis offered by Athena Diagnostics ($575) is nearly $100 more than the average price of the same test offered by the other providers. Athena’s price is nonetheless in the middle of the price range for full-sequence analysis offered by universities, hospitals and academic medical centers ($290–$816). The price per amplicon for GJB2 sequence analysis offered by several non-profit providers (range $140.8– $430) is comparable to Athena’s price ($287.50)
The cost of the MTRNR1 test offered by Athena Diagnostics ($365) is higher than the price of the test offered by universities and hospitals ($150– $285, average price $210). Athena’s higher price is not necessarily due to patents, however, and other factors may also contribute to price difference.
Testing for the MTTS1gene, which is not patented, is offered at prices comparable (average price $238) to MTRNR1 by universities and hospital-based providers. The test is not offered by any commercial testing providers, including Athena Diagnostics.
The SoundGene™ diagnostic panel developed by Pediatrix includes testing for the most common mutations associated with hearing loss, including GJB2/Connexin 26. Athena Diagnostics has negotiated a sublicense with Pediatrix for Connexin 26 testing. A guaranteed royalty stream from high volume of testing associated with newborn screening follow-up was a likely motivator of this agreement.
Communication and Marketing
Patents on hearing loss genes and related genetic tests appear to have little to no impact on dissemination of information about genetic testing or on how tests are marketed.
Athena Diagnostics does not engage in direct-to-consumer marketing. Athena markets primarily through a sales force keyed to clinical specialists. Athena does not have a sales force dedicated to the marketing of hearing loss tests to pediatricians or hearing loss specialists, rather its sales representatives address many neurological and neuromuscular conditions.
Adoption by Clinical Providers
To date, exclusive US licenses to patents on Connexin 26 and MTRNR1 testing do not appear to have secured Athena Diagnostics sole provider status. While Athena Diagnostics is the reference provider, a number of additional providers, most of which are academic medical centers, are listed as providers of clinical testing at GeneTests.org. However, Athena has intermittently enforced its patents, and laboratories remain concerned about future enforcement activity.
Negative effects of patents and licensing practices on adoption of genetic tests for hearing loss by providers are not readily apparent, although concerns were expressed in interviews. As early as 1998, ten providers offered GJB2 testing. The number of providers listed on GeneTests.org has risen to 18 since then. Nine providers for MTRNR1 testing are listed on GeneTests.org. However, there has been intermittent enforcement, and some providers have ceased offering some patented tests. We cannot determine how many laboratories decided against offering tests in the first place due to concerns about patent enforcement.
Athena Diagnostics has sent at least three “cease and desist” letters to other providers. In one instance, the UCLA Diagnostic Molecular Pathology Laboratory (non-profit) stopped offering a test (GJB2 and GJB6 as part of the panel) and did not negotiate a sublicense, citing substantial up-front payment as an impediment. GeneDx (for-profit) continues to perform full sequence analysis for Connexin 26 to identify mutations associated with a rare skin condition, KID, and agreed not to report hearing loss-associated mutations that are discovered during its full sequence analysis. In August 2008, the Center for Human Genetics of Boston University agreed to stop offering GJB2 testing, along with many other tests for which Athena Diagnostics holds an exclusive license.6
Providers of GJB2 and MTRNR1 testing presumably either collect samples and send them to Athena or offer the service without a sublicense from Athena Diagnostics. Athena states that no sublicenses for hearing loss testing have been negotiated with universities or academic medical centers to date.
Consumer Utilization
We found no evidence that consumer utilization of these tests is impeded by patents.
A large number of providers offer these tests with a wide price range.
Athena Diagnostics does not engage in direct to consumer marketing. There is no evidence that tests may be over utilized by consumers.
Given the lack of clear correlation between the costs of these tests and patent or license status, there is no evidence that patenting or licensing has hindered consumer utilization in the U.S. because of test price.
Some consumers (such as those covered by MediCal) may not have tests such as Connexin 26 testing covered by their insurance or health plan, because the reference provider Athena Diagnostics does not have a contract with that program. Access for these consumers therefore depends on the availability of additional providers who may have contracts with Medicaid or entails direct out-of-pocket payment by consumers. Uncertainty surrounding whether these alternate providers will face enforcement or will stop testing creates an unstable situation.
Adoption by third party payers
In our informal phone survey, test providers indicated that genetic tests for hearing loss are usually covered by insurance.
While comprehensive data on the coverage position of all major insurers for all hearing loss tests are not available, it is unlikely that patents have had significant impact on the adoption of tests. CIGNA health care, for example, covers testing for GJB2 (patented) and GJB6 (unpatented).
CLINICAL AND SCIENTIFIC BACKGROUND
Hearing loss refers to the permanent, bilateral or unilateral, sensory or conductive, loss of hearing averaging 30 decibels or more in the frequency region important for speech recognition.3 Hearing loss can present at different stages in life, and therefore can be classified as prelingual (before learning to speak) or postlingual (after having learned a language). Prelingual hearing loss may be congenital or late-onset. Profound congenital hearing loss occurs in 1.8 per 1,000 live births in the U.S. The prevalence increases to 2.7 per 1,000 among those below five years of age. During the teenage years, prevalence increases to 3.5 per 1,000.7 The lifetime societal costs for childhood hearing loss are estimated at $1.1 million per person, including lost productivity, special education, vocational rehabilitation, medical costs, and assistive devices attributable to deafness. Universal audiological newborn hearing screening programs have been introduced in the U.S. to reduce speech, social and emotional development problems experienced by children through early detection and intervention. At least 37 states have universal newborn hearing screening legislation and every state has early hearing detection and intervention programs, which screen approximately 93% of all infants.
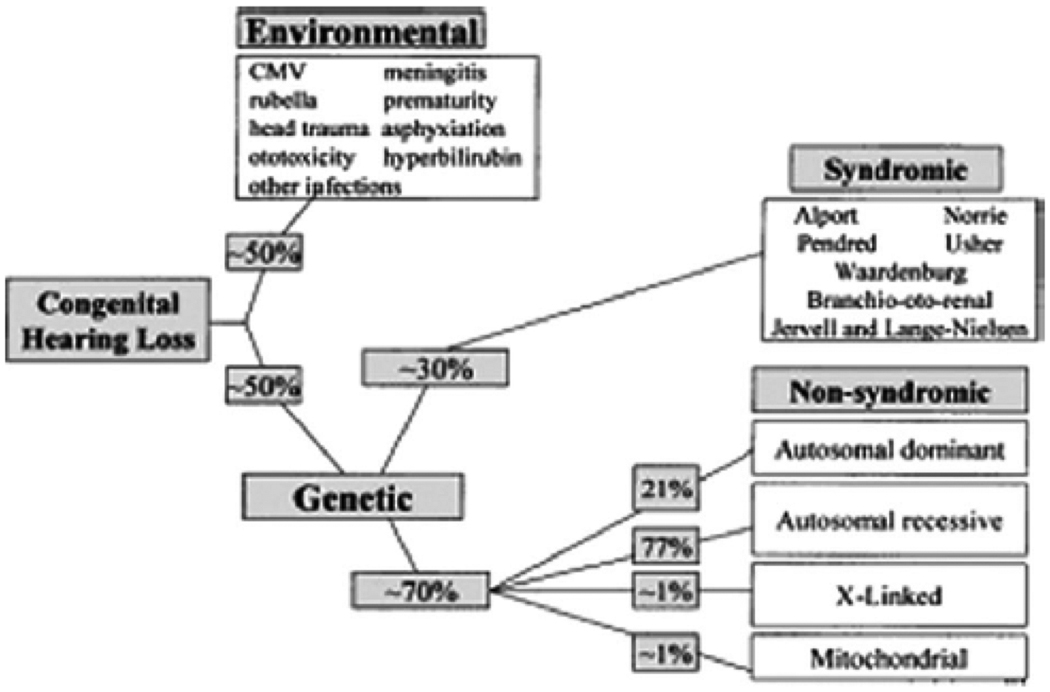
As a heterogeneous trait, hearing loss has many environmental and genetic causes. Its incidence varies over time and across populations (see Figure 1).7 Environmental causes, such as infections, account for approximately half of hearing loss cases. Congenital cytomegalovirus (CMV) infection, in particular, is responsible for as much as 10 percent of congenital hearing loss.7 Genetic causes account for the other half of hearing loss cases. Hearing loss typically occurs due to abnormalities in single genes or sometimes gene pairs. A multitude of different genes and gene pairs (at least 65 genes and 110 choromosomal locations) have been implicated. Many others may yet be discovered.7
Figure 1.
Causes of Hearing Loss (White K. Early hearing detection and intervention programs: opportunities for genetic services. Am J Med Genet 2004. 130A(1):29–36.) Copyright © 2004, John Wiley & Sons, Inc. Reproduced with permission of John Wiley & Sons, Inc.
Genetic hearing loss can be further classified as “syndromic” and “non-syndromic,” depending on whether it is associated with other clinical features (syndromic) or not (non-syndromic).7 Syndromic cases represent about 30 percent of genetic hearing loss cases overall and encompass at least 400 syndromes and a similar number of genes. Non-syndromic hearing loss or impairment (NSHL or NSHI) comprises the other 70 percent of genetic hearing loss cases and involves at least 100 loci, which can further be broken down by pattern of inheritance. NSHL loci include 55 autosomal recessive (AR), 41 autosomal dominant (AD), 4 X-linked (XL), and two mitochondrial loci. Different mutations at the same locus (chromosomal location, usually a gene) can present as either non-syndromic or syndromic hearing loss.3 Mutations in different genes may also result in the similar phenotypes (clinical symptoms and signs).7 A listing of non-syndromic and syndromic hearing loss disorders and loci, including genes, genetic tests, and associated patents, is presented in the appendix (Appendix I and II).
FIVE MOST COMMON GENETIC TESTS FOR HEARING LOSS
Given the numerous hearing loss genes, we have chosen to focus on the five genes that are most commonly tested for and the most common hearing loss genes: GJB2/Connexin 26, GJB6/Connexin 30, SLC26A4/PDS, MTRNR1, and MTTS1 (Personal communications with Dr. Michael Watson, Director, American College of Medical Genetics).
GJB2
Mutations in GJB2, or Gap Junction Protein Beta-2, have by far the highest frequency among genetic causes of deafness and hearing loss, accounting for up to 50 percent of cases of profound deafness caused by DNA mutations (Table 1).3 GJB2 encodes Connexin 26 (Cx26), a hexameric gap junction protein widely expressed in the cells and tissues of the cochlea.7 The link between GJB2 and non-syndromic deafness at the DFNB1 locus was first published in a 1997 Nature article by D.P. Kelsell and colleagues at St. Bartholomew’s and the Royal London School of Medicine and Dentistry, Queen Mary and Westfield College.8 (Nonsyndromic hearing loss loci are classified “DFNB” for recessive, “DFNA” for dominant, “DFN” for X-linked, and “DFNM” if they modify the expression of other genetic forms. The loci within each class are then numbered.7) That same year at the Institut Pasteur, Christine Petit and colleagues discovered the most prevalent GJB2 mutation, 35delG.9 The Institut Pasteur holds two patents (US 5,998,147 and 6,485,908) for the GJB2/Connexin 26 gene and detection of its common deletion mutation. Patent applications were filed in August 1997 and granted in 1999 and 2002. The Institut Pasteur also holds patents for Connexin 26 in Canada and Japan. We have found no granted patents in Europe, although applications appear to have been filed. Patents have been exclusively licensed to Athena Diagnostics, and we infer these were licensed for use in the U.S., Canada, and Japan. In Europe, the exclusive license for Connexin 26 testing went to Nanogen, a provider of molecular diagnostic services.10 As of February 2008, “Molecular Diagnostics for Prelingual Hearing” was still listed as a diagnostic technology available for licensing at the Institut Pasteur technology transfer website. This suggests either that existing licenses to Nanogen and Athena do not exhaust all territories worldwide or that provisions for particular fields of use have been retained by Institut Pasteur. As of January 2009, the technology is listed under Genomics (ID 98.30); however, it is unclear if the technology listed relates to testing for GJB2 specifically. Previous versions of the site accessed in February 2008 indicated that the technology listed was GJB2 testing.11 We have no direct information about whether Institut Pasteur has granted any additional licenses in Europe or the US. The Institut Pasteur was contacted by e-mail to clarify the status of licenses but has not responded.
TABLE 1.
Prices of Genetic Tests for the Five most Commonly Tested Hearing Loss Genes
| Gene Name |
Type of hearing loss |
Prevalen ce in affected |
Patent holder |
Type of Test |
No of providersa | Price of Test ($)b | ||
|---|---|---|---|---|---|---|---|---|
| NonPr ofit |
For Profit |
NonPr ofit |
For Profit |
|||||
| GJB2 | Non syndromic | >50%i | Institut Pasteur US 5998147 US 6485908 | Full sequence Analysis | 181 | 472.35c Athena (362–818) Preventiond Diagnostics | 575 290 |
|
| GJB6 | Non syndromic | 7–16%j | N/A | Deletion Analysis | 61 | 300.25e 295 (161–534) | ||
| SLC26A4 | Syndromic | 4–10%k | N/A | Full sequence Analysis | 60 | 1686f N/A (1100–2507) | ||
| MTRNR1 | Mitochondrial Non syndromic | <1%i | Cedars-Sinai US 5506101 | Targeted mutation | 82 | 210g h (150–285) | 248 365 |
|
| MTTS1 | Mitochondrial Non syndromic | <1%i | N/A | Targeted mutation | 40 | 238 N/A (150–285) | ||
Providers for specific test type identified from Genetests (see http://www.genetests.org) are current as of January 2009.
List prices of tests obtained from phone survey March 2007 or test laboratory web site
Average list price for 14 out of 17 providers offering full sequence analysis
Prices of 2 separate for-profit providers in 2008. Preventions Diagnostics is no longer listed on GeneTests as of January 2009.
Average list price for 4 out of 7 providers
Average list price for 4 out of 6 providers, not including NIH which offers testing free of charge to research participants
Average list price for 6 out of 8 providers
Prices of 2 separate for-profit providers
Based on data gathered through our telephone survey of providers (identified through GeneTests.org), testing for GJB2/Connexin 26 in the United States began as early as 1998. Kenneson et al. surveyed Connexin 26 testing providers in the U.S. in 1999 and 2000 (10 eligible providers in 1999 and 8 providers in 2000).12 Based on provider information at GeneTests.org, 19 U.S. providers (18 non-profit and 1 for-profit) offered full sequence analysis, which is the most common type of GJB2 testing. PCR-based sequence analysis has been facilitated by the relatively small size of the single GJB2 coding exon.13 Full sequence analysis is appropriate given that more than 195 GJB2 mutations have been identified,7, 14 which vary in frequency by race/ethnicity and family history.13 The average price of the GJB2 full sequence test among non-profit providers is $472.35 compared to the list prices of $575 quoted by Athena Diagnostics, the reference provider (Table 1).
Prices for full sequence analysis of Connexin 26, when normalized for number of amplicons, are also quite variable among providers. The unit price for the test offered by Athena Diagnostics is in the middle of the price range of non-profit providers (Table 2). The average price per amplicon of tests offered by non-profit providers is ~$231 and is comparable to Athena’s unit price for full-sequence analysis ($287.5). Although diagnostic billing codes provide some standardization for full-sequence tests, techniques and procedures are not identical among laboratories. The same billing codes are not always used, and the labs surveyed also likely have different overhead costs.
TABLE 2.
Comparison of Prices for Connexin 26 full Sequence Analysis
| Laboratory | Amplico ns* |
Gene sequencing price |
Cost per amplicon** |
|---|---|---|---|
| Athena Diagnostics (for profit) | 2 | $575 | $287.5 |
| Case Western University | 5 | $704 | $140.8 |
| Emory University | 3 | $490 | $163.33 |
| Univ of Chicago | 2 | $430 | $215. |
| Cincinnati Childrens Medical Center | 2 | $533 | $266.50 |
| Baylor College | 1 | $430 | $430 |
| Harvard Partners | 2 | $400 | $200 |
| Greenwood Genetics | 2 | $500 | $250 |
| Univ of Washington | 2 | $362.54 | $181.27 |
Number of nucleic acid sequences targeted for amplification (according to number of times CPT billing code 83898 is used)
Gene sequencing price divided by number of times CPT 83898 billed
While it appears that the number of U.S. providers offering Connexin 26 testing has increased to 19 from the 10 identified by Kenneson et al. in 2000 (19 providers listed on GeneTests.org offered full sequence analysis in January 2009),12 it is unclear whether the Institut Pasteur’s exclusive license to Athena Diagnostics for Connexin 26 testing has deterred other laboratories from testing. Some listed services may send samples to Athena or to offshore providers. To date, it appears that Athena Diagnostics has not granted sublicenses to any other providers listed on GeneTests.org (Personal communications with Dr. Michael Henry, Business Development, Athena Diagnostics). It is also not clear whether patents and exclusive licensing have contributed to a pricing differential or monopoly pricing by a sole provider. The 14 non-profit institutions we surveyed offer the test at varying prices, some comparable to the price of Athena Diagnostics, as shown in Tables 1 and 2.
GJB6
A significant portion (30–50%) of nonsyndromic genetic hearing loss is attributed to mutations in GJB6, or Gap Junction Protein Beta-6. Like GJB2, GJB6 is expressed in the cochlea and contributes to DFNB1 hearing loss. The GJB6 gene encodes Connexin 30 (Cx 30), a heteromeric gap junction protein that can form channels with Connexin 26, resulting in cases of digenic transmission (that is, the condition results from two different affected genes).7 The link between the > 300kb GJB6 deletion and nonsyndromic DFNB1 hearing loss was first published in January 2002 in the New England Journal of Medicine by Ignacio del Castillo and colleagues at the Unidad de Genética Molecular, Hospital Ramón y Cajal, Madrid, Spain.15 Genetic testing for GJB6 deletions in patients with hearing loss is linked to the genetic diagnosis of GJB2. GJB6 deletions are found in trans (that is, the genes are located on different chromosomes, suggesting the effect is mediated by a protein produced by the genes, rather than regulation of the genes themselves) with a mutant GJB2 allele and contribute to the same subtype of genetic deafness, DFNB1. The joint contribution of mutations in these two genes to non-syndromic recessive hearing loss is about 30–50%. While prevalence varies across populations, one North American study found a 2.57% prevalence of GJB2/GJB6 digenic cases among deaf individuals, with more severe hearing loss than is typical for GJB2 alone.13 However a more recent study by Putcha et al. reported that the frequency of a >300Kb deletion in individuals bearing compound GJB2 and GJB6 mutations was only 1% in a large North American cohort. Putcha et al.’s data suggest that this mutation may be quite rare.16 No U.S. patents or applications associated with the Connexin 30 gene or mutation testing were identified in our patent searches. Dr. Ignacio del Castillo, who first reported the GJB6 deletion mutation, confirmed that he had not applied for patents (Personal communication with Dr. Ignacio del Castillo). To date, seven (six non-profit and one for-profit) providers offer Connexin 30 deletion analysis in the U.S. The test appears to have been offered in the U.S. as early as 2002, based on our telephone survey of providers listed on GeneTests.org. The list price for GJB6/Connexin 30 testing averages $300 at non-profit institutions and is $295 at the one for-profit laboratory.
SLC26A4
In 1997, Eric Green and colleagues at the National Human Genome Research Institute (NHGRI) identified the SLC26A4, or PDS gene, which encodes the protein pendrin, a transporter of chloride, bicarbonate and iodide.17 Mutations in SLC26A4 are implicated in a form of syndromic deafness (Pendred syndrome), as well as a form of nonysndromic deafness DFNB4. Pendred syndrome is the most common form of syndromic deafness, and accounts for up to 10 percent of deafness. Pendred syndrome has an incidence of 7.15–10 per 100,000 births.17
While both Pendred syndrome and DFNB4 involve severe hearing loss and an enlarged vestibular aqueduct, Pendred syndrome is also associated with thyroid goiter. In the absence of a goiter, Pendred syndrome is classified by an abnormal perchlorate discharge test.18 The severity of goiter is variable, and thyroid symptoms may not occur until late childhood or even adolescence. Pendred syndrome typically has a prelingual age of onset (before the critical period for language development), whereas nonsyndromic DFNB4-associated deafness tends to be postlingual.7 No U.S. patents relating to SLC26A4 were identified.
Based on our informal phone survey of providers, testing for SLC26A4 has been available since at least 2002. The most commonly offered test, full-sequence analysis, can detect disease-causing mutations in about half of multiplex and one-fifth of simplex cases.18 All six U.S. providers of full sequence analysis SLC26A4 testing are non-profit institutions, and the average price is $1,686. The relatively high price of SLC26A4 full sequence analysis cannot be attributed to the existence of a patent or exclusive licensing. Rather, it appears that the cost of full sequence analysis relates to SLC26A4 being a large gene (~77 Kb) with twenty-one exons encoding a 4.93 Kb transcript. Therefore, testing requires testing methods comparable in complexity and price to testing for inherited susceptibility to colon and breast cancer.7, 19 The price/per amplicon for sequencing the SLC26A4 gene ranges from $55.00– $125.25 when standardized for the number of PCR amplifications reactions performed. The number of amplicons for SLC26A4 gene sequencing is 20, which is the number of nucleic acid sequences targeted for amplification (based on the number of times CPT billing code 83898 is used by the provider). Four providers offer SLC26A4 analysis for specific mutations at lower costs ($635) than the full sequence analysis. Targeted mutation analysis has a sensitivity of 70% for heterozygotes and 91% for those homozygous for a mutation.7
MTRNR1 and MTTS1
Mitochondrial forms of moderate to profound nonsyndromic hearing loss result from mutations in either the MTRNR1 or MTTS1 genes in mitochondrial DNA, each of which accounts for fewer than 1% of hearing loss cases. MTRNR1 encodes 12S ribosomal RNA (12S rRNA), while MTTS1 encodes transfer RNA for serine (tRNA Ser[UCN]).20 The most common MTRNR1 mutation, A1555G, occurs with a 0.3% frequency in the United States.20 Prezant et al., from Cedars-Sinai Medical Center in Los Angeles, California, first reported the association between A1555G mutations and aminoglycoside-induced and nonsyndromic deafness in Nature Genetics in July 1993.21, 22
MTRNR1 mutations may contribute to permanent, non-progressive hearing loss either through: (1) susceptibility to aminoglycoside (antibiotic) ototoxicity, irrespective of dose, or (2) late onset hearing loss in the absence of aminoglycoside exposure. MTTS1-related hearing loss, in contrast, has a characteristic progression first appearing during childhood and with penetrance that varies by individual mutational load (more numerous mutations accompany earlier onset and more severe deafness).20 Higher mutation loads of some MTTS1 mutations also correlate with the manifestation of other clinical signs, such as palmoplantar keratoderma, or ataxia and myoclonus.
The association between mutations in MTTS1 (tRNA –Ser [UCN]) and sensorineural deafness was first reported in 1994 by F.M. Reid and colleagues at the University of Glasgow in Scotland, UK.23
Cedars-Sinai Medical Center holds a patent (US 5,506,101) that covers MTRNR1 mutation testing, specifically testing for the A1555G mutation. The patent application was filed in June 1993 and granted in April 1996. Athena Diagnostics acquired an exclusive license for mutation testing for MTRNR1 from Cedars-Sinai Medical Center. Cedars-Sinai Medical Center also holds patents in Japan and Canada for MTRNR1 A1555G mutation and testing. No patents were filed in Europe.
Our searches found no patents covering the MTTS1 sequence or genetic testing for its mutations.
MTRNR1 testing first became available in the U.S in 2000. Targeted mutational analysis is now offered by ten U.S. providers. The two for-profit providers average a higher list price ($355) than the six non-profit (university hospitals and medical center based) providers (average $210) (See Table 1). Information about sublicenses from Athena Diagnostics for MTRNR1 mutation testing is not publicly available. (If SACGHS sends a list of queries to Athena Diagnostics, licensing status of MTRNR1 testing could be included.) In contrast, MTTS1 targeted mutation analysis has been available since 2004 and is offered by four non-profit providers for an average price of $238 (see Table 1). In addition, a subset of non-profit providers also offers testing for a panel of mitochondrial mutations, including both MTRNR1 and MTTS1, for an average price of $438.
NEWBORN HEARING SCREENING
Because of the potential for language, social, emotional, and other developmental consequences in children whose hearing loss is detected after six months of age, a 1993 National Institutes of Health (NIH) Consensus Development Conference endorsed universal newborn hearing screening.24 In 1999, the Health Resources and Services Administration (HRSA) and Centers for Disease Control and Prevention (CDC) began funding state Early Hearing Detection and Intervention (EHDI) programs.25 At least 37 states have legislation for universal newborn screening for hearing. Today, EHDI programs exist in every state, providing screening for approximately 93% of all infants.26 The goals of EHDI programs are three-fold: (1) to screen all newborns before one month; (2) to diagnose newborns before three months; and (3) to coordinate intervention before six months (see Appendix III for detailed flowchart).25 EHDI programs have reduced the average age for confirming hearing loss from 20 to 30 months (before the program), to 2 to 3 months (after implementation).
The EHDI programs miss some hearing loss cases, however, because prelingual hearing loss does not always present during infancy. SLC26A4 and A1555G-related hearing loss can appear after infancy, for example. Some cases of GJB2 deafness cannot be detected at birth. With an estimated non-penetrance rate of 3.8%,27 EHDI programs are seen by some as an opportunity for more genetic testing as part of the evaluation process.28, 29 Practical obstacles remain in screening programs for hearing loss, including uncertainty about the appropriate timing and role of genetic testing in the EHDI process.30 Survey data show that 20% of professionals who administer EHDI programs lack genetics training, which fuels concern about ordering and interpreting complex genetic tests.1
CLINICAL GUIDELINES FOR GENETIC TESTING
In 2002, the American College of Medical Genetics (ACMG) published clinical guidelines that incorporate genetic testing into the diagnosis of congenital hearing loss.3 The Cincinnati Children’s Hospital Medical Center’s testing paradigm exemplifies how hearing loss genetic test providers approach genetic evaluation (see Appendix IV). A pre-test session to explain the causes and types of deafness, along with testing options and modes of inheritance, is important. After the pre-test session, the next step entails getting a family history and an individual patient history and conducting a physical examination to determine whether or not a diagnosis is apparent. If syndromic hearing loss is suspected, the ACMG recommends gene-specific mutation tests. The diagnosis of non-syndromic cases is more complex, and relies on details of family history and individual symptoms. Individuals with hearing-impaired first-degree relatives, or two deaf parents, are also candidates for GJB2 testing. As the most common genetic cause of hearing loss, GJB2 is the first in a series of recommended tests.
If a GJB2 test reveals that an individual is a heterozygote, Cincinnati Children’s conducts a follow-up GJB6 deletion screen. If the GJB2 test is negative, the ACMG calls for non-syndromic mitochondrial testing, specifically for the A1555G and A7445G mutations. Cincinnati Children’s distinguishes among the types of mitochondrial testing, suggesting MTRNR1 testing only in the presence of aminoglycoside exposure, and a full mitochondrial panel otherwise. Following these initial rounds of genetic testing for GJB2 and mitochondrial mutations, the ACMG recommends post-test counseling and education. Given that 10% of deaf infants have culturally deaf parents, the availability of interpreters and the culturally sensitive interpretation of hearing loss test results are critical.3
After parents are informed of their options, follow-up and additional genetic testing may be recommended. Imaging studies may be ordered to consider the possibility of DFNB4 or Pendred syndrome, particularly for progressive hearing loss. Such imaging studies may include temporal bone imaging, to look for an enlarged vestibular aqueduct and/or cochlear dysplasia. If imaging studies have positive findings, mutation screening of SLC26A4 would be recommended.
Clinical Utility of Genetic Testing for Hearing Loss
Genetic tests offer several advantages over conventional hearing loss evaluation without genetic testing. The benefits anticipated from genetic testing include:29–31
Reduction of additional time consuming, invasive, and expensive testing;
Choice of early interventions such as hearing aids, cochlear implants, or sign language that significantly improve language ability and quality of life outcomes;
Information on the progression of the condition;
Ability to monitor associated clinical manifestations and complications, particularly for certain syndromic forms of hearing loss;
Information on the chance of recurrence in the family that can inform reproductive decisions; and
Information pertinent to risks and health care decisions (e.g., avoiding aminoglycoside antibiotics among those with MTRNR1 mutations).
Genetic testing may be more sensitive and specific than traditional evaluation. A study at Cincinnati Children’s Hospital found that 80% of hearing loss patients remained undiagnosed after traditional evaluation. Furthermore, genetic tests may facilitate earlier detection of hearing loss. Despite widespread newborn screening for hearing loss, a recent analysis showed that newborn screening can fail to identify some infants with two GJB2 mutations.13 The age at which the hearing loss was identified ranged from 12–60 months. A delay in detecting hearing loss has important implications for language acquisition and limits subsequent choices among management strategies. A study about cochlear implants reports, “There seems to be a substantial benefit for both speech and vocabulary outcomes when children receive their implant before the age of 2.5 years” (p. 628).32 A white paper addressing the societal costs of hearing loss concludes that “early identification of deafness or hearing loss is critical in preventing or ameliorating language delay or disorder in children who are deaf or hard of hearing and allows for appropriate intervention or rehabilitation. Early identification and intervention have lifelong implications for language development.”33 The present value of lifetime societal costs for prelingual hearing loss is estimated as $1.1 million, which includes lost productivity, special education, vocational rehabilitation, medical costs, and assistive devices attributable to deafness.34
Cost Effectiveness of Genetic Testing for Hearing Loss
We found no comprehensive cost effectiveness analyses of genetic testing for hearing loss. GJB2 testing may preclude the need for more expensive or invasive tests and provide the emotional benefit of knowing the cause as well as the clinical benefit of predictive information about progression and treatment options.28 A recent study at the Cincinnati Children’s Hospital Medical Center demonstrated that when compared to simultaneous testing, which comprises a battery of tests including standard laboratory work-up, or diagnostic evaluation by imaging, a diagnostic algorithm with GJB2 genetic testing as the first step resulted in a possible savings of “$20,180 in imaging costs and $34,000 in laboratory test costs per 100 children” screened (p. 809).35 The data on test-specific savings are in Table 3.35
TABLE 3.
Cost Estimates of Alternative Snhl Evaluation Approaches Based on Diagnostic Yields
| Testing yields |
Bilateral | Unilateral | Overall | ||
|---|---|---|---|---|---|
| Mild to moderate |
Moderately severe |
Severe to profound |
|||
| GJB2 screen (N = 161) | 15.5% (N = 45) | 5.0% (N = 20) | 37.7% (N = 71) | 0.0% (N = 25) | 18.0% |
| Imaging (N = 616) | 21.2% (N = 144) | 24.7% (N = 81) | 29.9% (N = 241) | 35.7% (N = 150) | 27.3% |
| Laboratory test | 0.0% | 0.0% | 0.3% | 0.0% | 0.07% |
| Cost estimates (per 100 children) | |||||
| Simultaneous evaluation | $193,200 | $193,200 | $193,200 | $193,200 | $193,200 |
| GJB2 paradigm | $141,096 | $152,005 | $121,530 | n/a | $139,020 |
| Imaging paradigm | $145,900 | $144,034 | $144,763 | $103,00 | $145,766 |
Reprinted from Otolaryngology – Head and Neck Surgery, Vol. 131, num. 6, Diego A. Preciado, Lynne H.Y. Lim, Aliza P. Cohen, Colm Madden, David Myer, Chris Ngo, John K. Bradshaw, Louise Lawson, Daniel I. Choo and John H. Greinwald Jr, “A diagnostic paradigm for childhood idiopathic sensorineural hearing loss,” pp. 804–809, at p. 809, Copyright 2004 American Academy of Otolaryngology – Head and Neck Surgery Foundation, Inc., with permission from Elsevier.
Another study at Children’s Hospital of Alabama assessed the cost of a battery of laboratory tests to evaluate hearing loss, including thyroid function, congenital infection, electrocardiograms, urine analysis, and serum phytanic acid levels, weighed in at more than $1,300, compared to the one-time $425 cost of a GJB2 genetic test.31
While the benefits of GJB2 testing have yet to be quantified, researchers note the ability of GJB2 tests to define chance of recurrence, i.e., if a child is GJB2 positive, a hearing couple knows that there is a 25 percent chance they will have a deaf child in each future pregnancy, and a deaf couple (each with GJB2 deafness) can learn that there is a 100% chance they will have deaf children.31 GJB2 testing may also be important given the success of cochlear implants among GJB2 positive individuals. A GJB2-positive individual may develop the same speech skills as an individual with normal hearing if the hearing loss is diagnosed and the cochlear implants are prescribed at a young enough age.31
In the case of non-syndromic mitochondrial testing, quantitative data are scarce. The benefits, however, are significant, considering that a positive A1555G test could prevent an infant from being exposed to aminoglycoside antibiotics, thereby preventing hearing loss. Another consideration associated with testing for these mutations is that aminoglycosides are often given before genetic testing has been performed because the infectious process has to be treated without delay. So in reality, the test is only beneficial if conducted prior to the onset of infection, or if test results can be turned around within a few hours. Due to increased numbers of premature births and widespread use of gentamycin in neonatal intensive care units, neonatologists have been particularly concerned about A1555G mutations and aminoglycoside exposure. However, in the absence of point-of-care testing, it would require screening parents prior to delivery or testing newborns to identify those at high risk of hearing loss from aminoglycoside use. For an individual with an A1555G substitution and no exposure to aminoglycosides, the probability of developing hearing loss by age 30 drops from 100% to 40%.13 Given the lifetime cost associated with prelingual hearing loss of $1.1 million, that amount could be averted by each case of deafness avoided. Since aminoglycosides are only prescribed in the event of severe in-hospital infections, the number of individuals prescribed aminoglycosides and estimates of the increased risk of untreated infection would have to be factored into any cost-effectiveness calculation.36
The limitations of genetic testing for hearing loss also have to be taken into account in cost effectiveness analysis. Since genetic deafness is population- and ethnicity-specific, relative frequencies should first be refined to best represent the population being studied. While GJB2 testing may confer large benefits for individuals who test positive, those benefits also have to be measured against the costs for individuals who test negative. Individuals who test negative for GJB2 mutations may have to undergo additional medical and/or genetic testing or may experience emotional difficulty when attempting to comprehend the meaning of the confusing and inconclusive test results.31
Molecular Testing for Hearing Loss: New Developments and Technologies
If recommendations to include genetic testing as part of expanded EHDI programs and clinical follow up of infants identified by universal newborn hearing loss screening are followed, then the volume of genetic testing for hearing loss could rise dramatically.29, 37 Testing for mutations associated with the most common forms of syndromic and non-syndromic hearing loss plus congenital CMV infection can determine the cause of hearing loss in most cases of congenital hearing loss. Preciado et al. conclude that introduction of genetic testing (specifically GJB2 testing) for hearing loss in the clinical evaluation paradigm is cost effective.35
Recently Pediatrix introduced genetic testing services for hearing loss. Pediatrix is one of the largest providers of newborn metabolic screening and newborn hearing loss screening services in the U.S. Pediatrix’s SoundGene™ Screening panel includes mutations associated with the most common forms of nonsyndromic and syndromic hearing loss. It also includes testing for common mutations in the mitochondrial MTTS1 gene, as well as testing for CMV infection (determined by measurement of copies of viral DNA, and therefore also, in essence, another genetic test). CMV infections account for up to 25% of congenital hearing loss caused by pathogenic agents. The SoundGene™ panel includes:
The SoundGene™ Screening Panel38
Connexin 26 (Cx26) GJB2 mutations
35delG 167delT
235delC M34T
Connexin 30 (Cx30) GJB6 large deletion
309 kb large deletion
Mitochondrial mutations
7445A>C (A7445C) 961T>C (T961C)
7445A>G (A7445G) 961T>G (T961G)
7444G>A (G7444A) 961 delT + C(n)ins
Pendred SLC26A4 mutations
L236P 1001+1G>A
E384G T416P
CMV DNA
The SoundGene™ Screening Panel was introduced in December 2006. The list price is $ 95.00 (Personal communication with Pediatrix). A U.S. patent application for the SoundGene™ Screening Panel is pending (Application U.S. 20040038266A filed in 2003, see Appendix V). SoundGene™ has also been trademarked. The test is described as a “quick and cost-effective alternative” and has an average turnaround time of 48 hours. Genetic counseling services for interpretation of test results and consultation are available through Pediatrix. Pediatrix has acquired a sublicense from Athena Diagnostics for testing of the Connexin 26 35delG mutation, which is included in the SoundGene™ panel. Pediatrix is the only provider to which Athena reports having issued a sublicense for Connexin 26 testing in the U.S. Although we do not have details of the licensing agreement and royalties, it is likely that the anticipation of high testing volume by Pediatrix as part of its newborn hearing loss screening services was an incentive for this agreement. Interestingly, however, the SoundGene™ panel does not include testing for the common A1555G mutation in the mitochondrial MTRNR1 gene (Patent no: US 5,506,101) that is also exclusively licensed to Athena Diagnostics.
High-throughput molecular diagnostics for hearing loss
With over 90 percent of newborns currently being screened for hearing loss and the potential for expanded EHDI programs to include molecular screening, genetic testing may shift to newer platform technologies for high-throughput genetic testing. Microarray-based genetic testing is being actively pursued as an efficient, reliable and potentially cost-effective tool when many mutations in a gene or numerous different genes must be tested. Hearing loss could be such a case. Since hundreds of loci are involved in the biology of hearing loss and additional genes and mutations may yet be discovered, microarray chips that can readily add new genes or mutations might help address both research and clinical needs. Microarray-based diagnostic testing for hearing loss might make it more flexible, less expensive, and more comprehensive while being as sensitive and specific as existing genetic tests.
Several groups report working on microarray-based diagnostic testing for hearing loss. Henrik Dahl and colleagues from the University of Melbourne and Children’s Royal Hospital in Australia have developed a hearing loss microarray that detects 15 common mutations in the Connexin 26/GJB2, SLC26A4, USH2A genes and mitochondrial 12S rRNA.39 This array-based chip was validated using DNA from 250 patients diagnosed with sensorineural hearing loss. It detected the mutations for which it was designed with 100% accuracy, and Siemering et al. report that no false positives or negatives were detected.39 Commercial development of the hearing loss biochip is suggested by U.S. patent application US20070009887A1, “Genotyping of deafness by oligonucleotide microarray analysis,” which was filed in November 2003, listing Victoria Siemering and Henrik Dahl as the inventors (Appendix V).
Another microarray diagnostic chip was recently reported by Iris Schrijver, Andres Metspalu, and colleagues in September 2006.40 Their diagnostic panel includes 198 mutations in 8 genes most commonly associated with non-syndromic sensorineural hearing loss. A patent application US20070134691A1 for this diagnostic has been filed by Schrijver and colleagues (Appendix V). The chip uses arrayed primer extension (APEX) technology, first developed by J. M. Shumaker and C.T. Caskey (Baylor College of Medicine, Houston Texas) and A. Metspalu (University of Tartu, Estonia).41, 42 Patents covering this technology, US 6,153,379 and US 7,001,722, were granted in 2000 and 2006.
The hearing loss chip tests for mutations in Connexin 26/GJB2, Connexin 30/GJB6, GJB3, GJA1, SLC26A4, SLC26A5, and mitochondrial 12S rRNA and tRNA Ser[UCN] and includes the commonly tested Connexin 26 35delG and A1555G MTRNR1 mutations, both of which are licensed exclusively to Athena Diagnostics. Currently this diagnostic assay is being offered on a “research only” basis at the Molecular Pathology Laboratory at Stanford University by Dr. Schrijver and colleagues (Personal communication with Iris Schrijver). Genetic testing for hearing loss using this diagnostic chip is being offered by Asper Biotech.43 Asper Biotech, located in Tartu, Estonia, was founded in 1999 with Dr. Andres Metspalu as its scientific advisor, and has expertise in developing and validating highly customized SNP/mutation screening assays. Asper Biotech also offers genetic testing services for diseases including cystic fibrosis, Usher Syndrome, retinitis pigmentosa, thalassemia, and a panel of genetic disorders common in the Ashkenazi Jewish population.44 Dr. Andres Metspalu at University of Tartu, Estonia, confirmed that the testing services offered by Asper Biotech are for research. The hearing loss test and other genetic tests offered by Asper Biotech are used by some academic medical centers and hospitals in the U.S in clinical research studies, often as part of collaborative projects (Personal communication with Dr. Andres Metspalu, University of Tartu, Estonia, and founder of Asper Biotech). It is not clear that any licenses have been negotiated by Asper Biotech with Institut Pasteur or Nanogen for the use of Connexin 26 mutation testing or with Cedars Sinai Medical Center for MTRNR1 mutation testing, but a license might not be required because they are not patented in Estonia. (Patent applications covering Connexin 26 and MTRNR1 mutations and diagnostic testing were never filed in Estonia.) Dr Metspalu confirmed that there is no patent protection for Connexin 26 and MTRNR1 mutations and testing in Estonia. However, he indicated that if Asper Biotech did decide to market the hearing loss test in the U.S., it would have to acquire sublicenses for all the relevant intellectual property and would have to factor royalty payments into its business plan. (We are not sure we concur with this judgment if the test itself were conducted in Estonia.)
Additional groups in the U.S. (shown in Appendix V) are exploring the use of kits and microarray diagnostics for high-throughput, comprehensive, and cost-effective molecular screening. Dr. John Greinwald and colleagues at the Cincinnati Children’s Hospital previously reported that a diagnostic paradigm incorporating genetic testing during clinical evaluation of hearing loss proved more cost effective than standard simultaneous laboratory work-up.35, 45 Dr. Greinwald’s group is now testing a microarray-based diagnostic gene chip that includes 13 genes associated with hearing loss. This collaborative project between Cincinnati Children's Hospital Medical Center and the University of Cincinnati Medical Center is being carried out at the Computational Medicine Center and is in an early phase of integrity and validation studies.46 Dr. Greinwald and colleagues have also filed U.S. patent applications US20050112598A1 and US20040166495A1, “Microarray-based diagnosis of pediatric hearing impairment-construction of a deafness gene chip,” based on the development of this gene chip (Appendix V). In a recent paper, Li et al. reported using a multiplex allele-specific PCR-based universal array (ASPUA), which combines Amplification Refractory Mutation System (ARMS) with array technology for clinical diagnostic testing of hearing loss mutations in parallel.47
Several groups have thus developed high-throughput diagnostic testing for hearing loss. U.S. patent applications filed by at least two of these groups on microarray-based gene chips suggest the potential for future commercialization of these diagnostic tests. However, we do not know if these tests will be adopted by clinical providers. Factors including test sensitivity, clinical utility and cost of the test are likely to significantly affect their uptake.
We also do not know whether the chip makers and testing service providers have licensed patents for mutations and methods associated with genetic tests for hearing loss. Neither have we studied whether use of short DNA probes on these chips would infringe existing patents, as this would require detailed analysis of claims and deep knowledge of the testing methods.
Finally, we note that full-genome sequencing technologies are progressing apace, and if such analysis became possible, then the basis for genetic testing would be individual genomic sequencing and comparing that sequence to known mutations associated with all genetic forms of hearing loss, rather than tests specifically keyed to hearing loss. The intellectual property implications are unclear, as they are for genetic testing of other clinical conditions.
LESSONS LEARNED ABOUT THE IMPACT OF PATENTS ON ACCESS TO HEARING LOSS TESTING
Research
We found no evidence about positive or negative effects of hearing loss gene patents on research in the field of hearing loss genetics. Basic research to determine the associations between candidate genes and their roles in various forms of hereditary hearing loss has steadily progressed. Research appears to be proceeding rapidly on rare forms of deafness that offer the prospect of a small market for diagnostic testing and are therefore unlikely to provide significant monetary incentives for genetic testing. Most genes associated with different forms of syndromic and non-syndromic deafness are not patented (Appendix I and II). Even among the five most commonly tested hearing loss genes, which are presumably of greatest commercial interest, three genes are not patented. It is unclear whether patents or the potential for commercialization provided an incentive for the research. At least two research groups at non-profit institutions were engaged in studies to identify Connexin 26 gene mutations. Publications reporting the identification of mutations in Connexin 26 by Kelsell at al (Queen Mary and Westfield College, UK) and Christine Petit et al (Institut Pasteur) were submitted in January (published in May) and August (published in November) of 1997 to Nature and Human Molecular Genetics, respectively. While the UK group does not appear to have applied for a patent, Christine Petit and Institut Pasteur secured US patents on GJB2/Connexin 26 and its mutations in December 1999. Petit and colleagues applied for a patent in August 1997, the same month they submitted their findings for publication. Dr. Fischel-Ghodsian and colleagues at Cedars-Sinai Medical Center submitted their report on the MTRNR1 A1555G mutation to Nature Genetics in February 1993 (published July 1993). The corresponding patent application on detection of A1555G mutation was filed on June 30, 1993, four months after submitting for publication, and granted to Cedars-Sinai in April 1996. While these chronologies suggest that scientific publication and patenting activities proceeded in parallel, we cannot determine if journal submissions were in fact delayed in the first place to prepare patent applications for parallel filing.
Without information on the royalties Institut Pasteur and Cedars-Sinai Medical Center receive from the licenses to Athena Diagnostics for Connexin 26 and MTRNR1 testing, it is also difficult to comment on the impact these patents have had on supporting subsequent basic research at these institutions. Such support would be one of the positive effects of patents.
A substantial amount of clinical research has been performed, for example on the prevalence of Connexin 26 mutations in different populations, and on new methods for diagnostic testing including array-based diagnostics. Such studies include genetic testing for mutations covered by patents and licensed exclusively to Athena Diagnostics (Connexin 26, MTRNR1). However, researchers at academic medical centers told the authors that they remain concerned about the consequences of future enforcement activity by Athena Diagnostics on the clinical testing and clinical research. Researchers warn that uncertainty about whether an academic medical center or reference lab may be required to stop testing and the absence of a clearly stated policy about research use from Athena Diagnostics may have chilling effects on clinical research.
Development and Commercialization
Genetic tests for Connexin 26 and MTRNR1 which are patented, and for GJB6, SLC24A6, and MTTS1, which are not covered by patents, have been developed and are offered by several providers at similar prices. Several providers have in fact developed test panels that include both the patented Connexin 26/MTRNR1 as well as the unpatented Connexin 30/MTTS1 tests. The acquisition of an exclusive license for Connexin 26 diagnostic testing in the US was presumably integral to Athena Diagnostics’ plan to commercialize these tests. GJB2 testing was offered by at least 9 providers in the U.S. as early as 1998. The number of providers listed at GeneTests.org has doubled since 1999–2000.12 Testing for the patented genes GJB2 and MTRNR1 and their most common mutations is offered by more U.S. providers than testing for the unpatented genes SLC26A4, GJB6, and MTTS1. This is not entirely surprising given that GJB2 mutations account for up to 50% of cases of non-syndromic hearing loss. The majority of laboratories listing the tests are academic health centers.
Clinical testing for MTRNR1 in the U.S. may have been delayed. The association of MTRNR1 mitochondrial mutations to hearing loss was published as early as 1993, yet clinical testing appears to have become available only in 2000. In our telephone survey, many laboratories were unable to provide data on when they first made this test available. A more systematic and detailed survey of providers might help determine if patents impeded or deterred providers from developing these tests, as we did not query providers specifically about this issue.
It is difficult to assess exactly how much of a price premium the exclusive license provides Athena Diagnostics, or what impact the patent licenses have on volume. According to Athena Diagnostics, to date only one sublicense for Connexin 26 testing has been granted (to Pediatrix). Thus, the list price of the other providers must not include royalty or licensing fees. The price range can be attributed to factors such as overhead costs at different institutions. In the case of testing for MTRNR1, the price offered by both for-profit providers is on average $145 more than the price of the test provided by non-profit institutions. The $365 list price of the test offered by Athena Diagnostics is nearly 73 percent higher than the average list price offered by other university and hospital-based providers. In contrast, testing for the unpatented MTTS1 gene is offered by only four non-profit providers and at prices comparable to MTRNR1 testing services offered by these providers. MTTS1 testing is not offered by Athena Diagnostics.
Costs of hearing loss tests do not appear to correlate strongly with patent status. For instance, the price of the most expensive test--SLC26A4 full sequence analysis--can be attributed mostly to the costs of sequencing a large gene. The relatively high cost of the SLC26A4 testing also affects fewer consumers, since Pendred’s syndrome accounts for a small fraction of hearing loss cases and testing is recommended only to follow up on positive imaging findings.
Communication/ Marketing
It appears that patents on DNA sequences and platforms for hearing loss genetic testing have had little impact on the dissemination of information about such tests or how they are marketed. We found no evidence of direct-to-consumer marketing. In the course of a phone conversation, Dr. Michael Henry, Vice President of Business Development at Athena Diagnostics, clearly stated the company’s commitment to refrain from direct-to-consumer marketing and emphasized that Athena relies primarily on physician-prescribed testing. He also indicated that while Athena Diagnostics does have sales representatives who communicate information about genetic testing for neurological conditions to neurologists and medical practices, there is no sales force specifically committed to marketing hearing loss genetic testing to pediatricians and specialists (e.g., otolaryngologists and audiologists).
Adoption by Clinical Providers and Testing Laboratories
Any effects of patents on adoption of hearing impairment genetic tests by clinical providers are not readily apparent.
The exclusive license procured by Athena Diagnostics for Connexin 26 and MTRNR1 testing does not appear to have established Athena Diagnostics as the sole provider. However, the number of providers currently available may not fully capture the effects of patents on provider adoption. According to Dr. Michael Watson, Director of the ACMG, “Athena aggressively enforced their IP for many years but were increasingly irritating practitioners and made them an example in the press of bad IP behavior. Around 2000, they [Athena] stopped enforcing and tried to develop their ‘Academic Partnership Program.’ Although the intent was to allow laboratories to retain some volume for research and training of clinical laboratorians, it ultimately failed largely because if a lab did more than 100 cases in a year, the licensing fees made the lab noncompetitive” (Personal communication with Dr. Michael Watson).
We have clearly identified three instances of patent enforcement by Athena Diagnostics for Connexin 26 testing against other providers. The first of these proved to be a case of non-infringing use that has been resolved (Personal communications with Richard Flaherty, CIO, Director of Technology and Investor Relations, BioReference Laboratories (BRLI), and Sherri Bale, Clinical Director, GeneDx). In testimony before the House Judiciary Subcommittee on Courts, the Internet and Intellectual Property, on October 30, 2007, Marc Grodman, CEO of BioReference Laboratories, indicated that while GeneDx (a company acquired by BRLI) was performing a genetic test for a rare skin condition by full sequence analysis of the gene in question, it “received a letter from another laboratory claiming that within the sequence being analyzed was another sequence associated with hearing loss” (p. 4).48 Athena Diagnostics’ letter indicated that since testing for this hearing loss gene was patented, performing the test might be an act of infringement. Attempts by GeneDx to perform the test by paying a royalty to the other company were unsuccessful. We have confirmed by personal communication with Dr. Grodman and Dr. Sherri Bale, Clinical Director at GeneDx, that the genetic test in question involved sequencing the Connexin 26 gene for mutations associated with a rare skin condition Keratitis Ichthyosis Deafness (KID). Dr. Bale confirmed that Athena Diagnostics sent a “cease and desist letter” and indicated that the matter has been resolved. “We accepted a letter from Athena that instructed us to not report the 35delG mutation. However, what we've done is: if we find the deletion, we call the referring MD, tell them the results and that we can't report them, and then suggest they redraw the patient and send the sample to Athena for testing” (Personal communication with Dr. Sherri Bale). This requires a second visit to the patient’s physician, another blood draw, and payment, this time to Athena Diagnostics, to repeat the GJB2 test. (See Appendix VI Letter from Sherri Bale, GeneDx to Athena Diagnostics.)
GeneDx currently continues to perform full sequence analysis for Connexin 26 to identify the GJB2 D50N mutation and other mutations associated with a rare skin condition KID, which is not covered by the patents licensed to Athena. We understand the matter reached amicable resolution with GeneDx agreeing not to report hearing loss mutations and referring to Athena if they are found (See Appendix VII). Athena Diagnostics, which holds the exclusive license to GJB2 mutation testing in the U.S., expressed willingness to grant sublicenses (Personal communication with Michael Henry). However, according to Dr Sherri Bale, Athena refused to grant a sublicense when GeneDx attempted to acquire one in the context of KID testing (Personal communication with Richard Flaherty and Sherri Bale). This case also raises concerns about withholding of useful clinical information and increased costs, as another blood draw and test by Athena would be required if GeneDx identified a potential hearing loss mutation in a sample sent to them for KID testing, although this is clinically unlikely.
In another instance, the Diagnostic Molecular Pathology Laboratory at the University of California Los Angeles stopped offering testing for Connexin 26/GJB2 over two years ago, after receiving a “cease and desist letter" from Athena Diagnostics. According to Dr. Wayne Grody, Director of the Laboratory, the terms of the sublicense offered by Athena Diagnostics “were unreasonable, with an upfront fee of $50,000 per year plus a significant per test fee” and not economically viable for the laboratory, given the relatively low volume of testing for hearing loss at UCLA (Personal communication with Dr. Wayne Grody). Attempts to negotiate terms of a sublicense were not successful. It is unclear to what extent cessation of testing at UCLA has affected patient access to hearing loss testing. Dr. Grody indicated that samples are now sent to Athena Diagnostics for clinical testing. His laboratory considered using an alternate test methodology, namely custom ASRs from Third Wave Technologies for Connexin 26 mutation testing. This method reportedly allows laboratories to avoid infringing the Connexin 26 patents licensed to Athena. It is unclear if this is because a sublicense acquired from Athena Diagnostics comes attached to the purchase of the ASRs or because the test methodology (Invader™ assay) offers “workarounds” of the patents (US5998147, 6485908). However, these ASRs are no longer being offered since HoloLogics Inc acquired Third Wave Technologies in June 2008.5
Dr. Grody indicated that even if the alternate methodology could help overcome the problem of patent infringement, it is not ideal because ASRs for the 235delC Connexin 26 mutation, found commonly in Asian populations, are not available from Third Wave. Testing for this mutation is particularly relevant at UCLA given the high Asian and Asian American population in California. Dr. Grody also noted that shipping samples to Athena Diagnostics is problematic for indigent patient populations covered by the California MediCaid program (MediCal). MediCal only reimburses laboratories with which it has a contract, which Athena does not have.
We also recently became aware that Athena Diagnostics sent a “cease and desist” letter to the Center for Human Genetics at Boston University regarding testing for a number of genetic conditions including hearing loss (Personal communication with Dr. Aubrey Milunsky, Director, Center for Human Genetics, Boston University). In August 2008, the Center for Human Genetics ceased testing for hearing loss and several other conditions for which Athena has exclusive IP rights.
Athena confirmed that no sublicenses have been given to university and academic or medical centers (Personal communication with Michael Henry).
The SoundGene™ panel offered by Pediatrix is performed under a sublicense from Athena Diagnostics for GJB2/Connexin 26 testing. To our knowledge, Pediatrix is the only provider that has received a sublicense from Athena Diagnostics to date. Presumably, this will lead to a steady royalty stream for Athena from genetic testing done by Pediatrix as part of newborn hearing loss screening, and a flow of patients for diagnostic follow up.
Microarray chip-based diagnostics for hearing loss are currently not available as a clinical service in the U.S. However, if chip based diagnostics do become commercialized, and if use of DNA probes on those microarrays infringe the patents that Athena has licensed, Athena Diagnostics could choose to demand a license for testing that includes patented sequences of Connexin 26 and MTRNR1. Simultaneous multi-gene testing also seems to be a departure from the current ACMG clinical guidelines, which call for a systematic utilization of genetic tests based on relative frequencies, family histories, patient symptoms and apparent diagnosis. Those guidelines might change, however, if microarray testing proved equally sensitive, specific, and accurate, while being faster and cheaper and identifying many mutations in different genes in a single test.
Consumer Utilization
This case study finds limited effects on patient access to genetic testing for hearing loss that can be directly attributed to patenting. The availability of genetic testing for hearing loss in California may be limited for MediCal patients because the patent-holder, Athena Diagnostics, lacks a contract with MediCal and is out-of-state. The issue here is not patents per se, but patents preventing other laboratories from offering the test under MediCal contract. The laboratories with MediCal contracts do not have sublicenses from Athena and Athena apparently does not have a contract with MediCal.
We were unable to identify systematic evidence beyond the MediCal situation noted above, that the patents have impeded utilization of hearing loss tests by people who are interested in or require testing. Testing for the genes licensed exclusively to Athena Diagnostics is not marketed directly to consumers by Athena or by other direct-to-consumer providers like DNAdirect. Sixteen providers other than Athena Diagnostics are listed on GeneTests.org as offering Connexin 26 testing. Nine providers in addition to Athena Diagnostics are listed for MTRNR1 testing. Many of these provider websites have detailed information on the availability and cost of both patented and unpatented hearing loss genetic tests. Although several providers for these tests have emerged, we found no information about usage of the tests by consumers.
While we did not query test providers about their testing volume or the number of patients requesting each test, a future survey could assess utilization of hearing loss tests by consumers. It would also be valuable to determine how frequently reimbursement for such tests is denied by insurers and payers, as coverage and reimbursement of genetic testing are likely to affect consumer use.
Finally, patient access may be affected, as much or more by factors other than patents, such as the lack of knowledge about the genetics of hearing loss, particularly among primary care physicians, and their low propensity to refer cases for genetic testing as follow-up.49 A recent survey by Duncan et al. noted that while 86% of pediatric otolaryngologists reported having easy access to genetic testing services for referral, many also identified “discomfort with various aspects of genetic testing” as a reason for not ordering genetic tests (p. 235).50 Lack of knowledge about genetic testing or about interpretation of test results may be a more significant barrier to test adoption by healthcare providers than patents.
Coverage and reimbursement by third party payers
We have no evidence that gene patents have directly affected third party payer coverage and reimbursement decisions for hearing loss tests. Laboratories report that insurers have generally adopted genetic testing for some hearing loss genes, as illustrated below by the coverage position from CIGNA HealthCare on “Genetic Testing for Congenital Profound Deafness.”
“CIGNA covers genetic testing for congenital, nonsyndromic, sensorineural, mild-to-profound deafness (DFNB1) as medically necessary for ANY of the following indications:
For diagnostic testing when the clinical examination and conventional studies suggest a diagnosis of congenital, nonsyndromic, sensorineural, mild-to-profound deafness (DFNB1)
- For carrier testing in EITHER of the following situations:
- when the individual has a first- or second-degree relative* with gene GJB2 or GJB6 mutation
- when the individual is the reproductive partner of a known carrier (deafness-causing mutation of gene GJB2 or GJB6) and the couple has the capacity and intention to reproduce
For prenatal testing when both parents are known carriers of deafness-causing mutation of gene GJB2 or GJB6 mutation.
CIGNA does not cover genetic screening for congenital, nonsyndromic, sensorineural, mild-to-profound deafness (DFNB1) in the general population because such screening is considered not medically necessary or of unproven benefit.
All individuals undergoing genetic testing for any reason should have both pre- and post-test genetic counseling with a physician or licensed or certified genetic counselor.”51
Aetna covers full sequence and targeted mutation analysis of GJB2/Connexin 26 and deletion analysis for GJB6/Connexin 30, but it excludes pre-implantation genetic diagnosis (PGD) for DFNB1, which is deemed an “unproven benefit at this time.” We have not verified whether other commercial insurers have a similar position, except through the interviews with testing laboratories. Indirect effects that patents may have on price might lead to a higher level of scrutiny by insurance providers if the tests are priced above other genetic tests, but hearing loss genetic test prices are in the same range as other case studies. Decisions about coverage for SLC26A4, MTRNR1 and MTTS1 may be case by case, because these conditions are not common enough to warrant an explicit coverage policy. These tests are likely handled similarly to tests for other rare conditions, covering tests in a routine price range and requiring special justification for expensive testing. During the informal phone survey, most test providers indicated that hearing loss genetic tests were mostly covered by insurance. However, we have no direct evidence about how often consumers are denied coverage for hearing loss testing, pay for them out of pocket, face high co-pay fees because of reimbursement limits, or encounter other factors that affect their choice to get such tests.
Athena Diagnostics has a policy of directly billing insurance providers for services when Athena is the contracted provider for that particular plan. However, when Athena is not a contracted provider and the insurer does not cover the testing in part or full, Athena guarantees as part of its Patient Protection Plan that “an eligible, enrolled patient’s liability will be limited to 20% of the cost of the test, even if the patient’s insurance plan pays nothing. (These programs are discussed at greater length in the spinocerebellar ataxia case study.) For patients enrolled in the Patient Protection Plan, any amount collected from the insurance company in excess of 80% of the amount billed will be refunded to the patient.”52 The Patient Protection Plan is not, however, available in all states, does not apply to government health programs (Medicare and Medicaid, for example) and does not apply to most insurers and health plans. Florida and Maryland are excluded, for example.
Athena Diagnostics does not participate in Medicaid but it does offer discounts to Medicaid patients through its financial assistance programs. If the test of interest is not covered by Medicare carriers, the patient will be required to pay for the test in advance. In such cases, if the Medicare carrier denies coverage of the test, the patient may have to pay the entire cost out of pocket, since Medicare patients are ineligible for Athena’s Patient Protection Plan. Thus insurance coverage, independent of the patenting status of the test, may limit patient access in some cases, specifically Medicaid patients, most Medicare patients, and those covered by health plans with which Athena does not have a contract. However, even in these cases, patients have the option of using other providers who may accept Medicaid, at least as long as those providers continue to offer the service.
CONCLUSION
Patents do not appear to have significantly impeded patient or clinical access for hearing loss genetic testing. Many institutions provide tests, even those covered by patents exclusively licensed to Athena Diagnostics, presumably without a sublicense. While Athena Diagnostics has sent out some “cease and desist” letters, enforcement is apparently incomplete, as several other testing services are listed on GeneTests.org. It is possible that the volume of testing at most institutions, even for Connexin 26, is not large enough to warrant more aggressive enforcement by Athena Diagnostics.
Given that experts have recommended incorporation of genetic tests into EHDI programs, use of genetic tests for hearing loss is likely to increase. The recent introduction of the SoundGene™ diagnostic panel by Pediatrix Screening is indicative of this trend. However concerns have been raised that a small panel such as SoundGene™ may not be ideal. For example, patients with GJB2 related hearing loss may be missed because they do not carry the mutations represented on the panel. More recent literature suggests it is not sufficient to test only the four common mutations associated with Pendred syndrome included in the panel. This is one reason many labs now sequence the entire SLC26A4 gene because targeted mutation testing misses many mutations. Multi-gene, chip-based tests may help address problems in diagnosing individuals who develop hearing loss as children or adolescents, and potentially reduce the cost and duration of diagnostic testing. These new diagnostics, although likely to detect a much broader range of mutations and gene variants, may also miss rare and novel mutations, especially for genes like GJB2 and SLC26A4, as patients often have new or private mutations. The clinical utility and analytical validity of such array-based tests also needs to be demonstrated. It remains to be seen whether patents on genes and mutations for hearing loss will impede development of multi-allele methods.
This case study illustrates the complexity of assessing the impact of patents on access to genetic testing. This is in part because of the number of genes and mutations involved, but also depends on patents and their enforcement. Aggressive patent enforcement might reduce the number of outlets for genetic testing, and for those not covered by health plans covering payment to Athena Diagnostics, this would reduce access. It therefore appears that access depends on an unstable intellectual property regime and the vicissitudes of payment contracts between health insurers and health care plans, on one hand, and different testing labs, on the other.
Genetic testing for hearing loss also illustrates several other features of intellectual property and genetic testing. Most of the patents for commonly tested genes are owned by academic institutions and licensed to Athena Diagnostics. The patenting and licensing practices of academic institutions are therefore linked to both the benefits and problems associated with having a single major provider. The case also illustrates the penumbra effect of exclusive rights to some mutations leveraging testing for others, although it is also clear from this case that the effect is incomplete since multiple providers are offering tests.
ACKNOWLEDGEMENTS
This case study was reviewed by Richard L. Faherty, Michael Henry, Wayne Grody, Iris Schrijver, Sherri Bale, Michael Hopkins, Ignacio del Castillo[0], and Michael Watson for the Secretary’s Advisory Committee on Genetics, Health, and Society.
FUNDING: This case study was carried out under grant P50 003391, co-funded by the National Human Genome Research Institute and US Department of Energy, and supplemented by funding from The Duke Endowment. The case study authors have no consultancies, stock ownership, grants, or equity interests that would create financial conflicts of interest. The Center for Genome Ethics, Law & Policy accepts no industry funding. Dr. Robert Cook-Deegan is listed on the British Medical Journal roster of physicians who have pledged to remain independent of industry funding <http://www.tseed.com/pdfs/bmj.pdf>; more details about how the case studies were done are noted in a 29 July 2009 letter to the Secretary’s Advisory Committee on Genetics, Health, and Society <http://www.genome.duke.edu/centers/gelp/documents/SACGHSResponsetopubliccomments.pdf>.
APPENDIX I: NONSYNDROMIC LOCI: KNOWN GENES, GENETIC TESTS, AND PATENTS
| Locus | Pattern of Inheritan ce |
Genes | Age of Onset |
Relative Frequency | Test Available | Patent Holder |
|---|---|---|---|---|---|---|
| DFNB61 | AR | PRES (SLC26A5) | higher among Caucasians | Northwestern (6602992) | ||
| DFNB1 | AR | GJB2 (Cx 26), GJB6 (Cx 30) | Prelingual | Up to 50% | GJB2 (Cx 26), GJB6 (Cx 30) | Institut Pasteur (5998147, 6485908) |
| DFNB2 | AR | MYO7A | Prelingual | MY07A | ||
| DFNB3 | AR | MYO15A | Prelingual | 2% incidence in Benkala, Bali | ||
| DFNB4 | AR | SLC26A4 | Postlingual | 4–10% | SLC26A4 | |
| DFNB6 | AR | TMIE | Prelingual | |||
| DFNB7/11 | AR | TMC1 | Prelingual | |||
| DFNB8/10 | AR | TMPRSS3 | DFNB8-Prelingual, DFNB10-Postlingual | |||
| DFNB9 | AR | OTOF | Prelingual | OTOF | ||
| DFNB112 | AR | CDH23 | ||||
| DFNB16 | AR | STRC | Postlingual | |||
| DFNB18 | AR | USH1C | Prelingual | |||
| DFNB21 | AR | TECTA | Postlingual | TECTA | ||
| DFNB22 | AR | OTOA | Prelingual | |||
| DFNB23 | AR | PCDH15 | Prelingual | |||
| DFNB28 | AR | TRIOBP | Prelingual | |||
| DFNB29 | AR | CLDN14 | Prelingual | |||
| DFNB30 | AR | MYO3A | Prelingual | |||
| DFNB31 | AR | WHRN | Prelingual | |||
| DFNB36 | AR | ESPN | Prelingual | |||
| DFNB37 | AR | MYO6 | Prelingual | |||
| DFNB67DFNB59 | AR | TMHSDFNB59 (pejvakin) | ||||
| DFNA1 | AD | DIAPH1 | Postlingual | |||
| DFNA2 | AD | GJB3 (Cx 31), KCNQ4 | Postlingual | KQCN4 | NeuroSearch A/S (6794161) | |
| DFNA3 | AD | GJB2 (Cx 26), GJB6 (Cx 30) | Prelingual | GJB2 >50% | GJB2 (Cx 26), GJB6 (Cx 30) | Institut Pasteur (5998147, 6485908) |
| DFNA4 | AD | MYH14 | Varies | 1% | ||
| DFNA5 | AD | DFNA5 | Postlingual | |||
| DFNA6/14/38 | AD | WFS1 | Prelingual | WFS1 | Washington University School of Medicine (WOO18787A1) | |
| DFNA8/12 | AD | TECTA | Pre or postlingual | TECTA | ||
| DFNA9 | AD | COCH | Postlingual | COCH5B2 | Brigham and Women's Hospital (7030235), Brigham and Women's Hospital & U-Antwerp (6730475) | |
| DFNA10 | AD | EYA4 | Postlingual | EYA4 | ||
| DFNA11 | AD | MYO7A | Postlingual | |||
| DFNA13 | AD | COL11A2 | Postlingual | COL11A2 | ||
| DFNA15 | AD | POU4F3 | Postlingual | |||
| DFNA17 | AD | MYH9 | Postlingual | MYH9 | ||
| DFNA20/26 | AD | ACTG1 | Postlingual | |||
| DFNA22 | AD | MY06O | Postlingual | |||
| DFNA28 | AD | TFCP2L3 | Postlingual | |||
| DFNA36 | AD | TMC1 | Postlingual | |||
| Wash U. School of Medicine (WOO18787A1) | ||||||
| DFNA44 | AD | CCDC50 | Postlingual | |||
| DFNA48 | AD | MYO1A | Postlingual | |||
| None Listed | AD | CRYM | Prelingual | |||
| DFN3 | XL | POU3F4 | Prelingual | |||
| Aminoglycoside Ototoxicity | Mitochondrial | MTRNR-1 (A1555G), MTTS-1 | Prelingual | A1555G <1% (1/20–40,000 births) | MTRNR-1, MTTS-1 | Cedars-Sinai (5506101) MTRNR1 |
| None Listed | TDC-1, TDC-2 | TDC-1, TDC-2 | Griffith, Kurima, Wilcox & Friedman (20040249139A1) | |||
| Dentinogenesis imperfecta type II (DGI-II) | DSPP | DSPP | Kong, Xiao, Zhao, Yu & Hu (2003018020A1) |
APPENDIX II: SYNDROMIC DISORDERS: KNOWN GENES, GENETIC TESTS, AND PATENTS
| Disorder | Type | Pattern of Inheritance |
Genes | Age of Onset |
Relative Frequency |
Prevale nce |
Test Availab le? |
Patent Holder (Patent Number) |
|---|---|---|---|---|---|---|---|---|
| Pendred's Syndrome | Syndromic | AR | SLC26A4 | Prelingual | 4–10% | SLC26A4 | ||
| Type 4 Barter'st Syndrome | Syndromic | AR or digenic | BSND, CLCNKA, CLCNKB | con-sanguineous Middle Easterners | ||||
| Branchio-oto-renal (BOR) Syndrome | Syndromic | AD | EYA1, SIX1 | 1 in 40,000 | EYA1, SIX1 | |||
| Alport Syndrome | Syndromic | AD | MYH9, COL4A5, COL4A3, COL4A4 | Rare | MYH9, COL4A5, COL4A3, COL4A4 | |||
| Fechtner's Syndrome | Syndromic | AD | MYH9 | Rare | ||||
| Sebastian Syndrome | Syndromic | AD | MYH9 | Rare | ||||
| (DFNA22) | Syndromic | AD | MYO6 | Postlingual | Rare | |||
| Renal Tubular Acidosis | Syndromic | AR, consanguinity | ATP6B1, ATP6N1B | con-sanguineous North Africans | ||||
| Waardenburg's Syndrome | Syndromic | AD or AR | PAX3, MITF, SOX10, EDN3, EDNRB | 1–4% | ||||
| Wolfram Syndrome | Syndromic | AD | WFS1 | Prelingual | WFS1 | Washington University School of Medicine (WOO18787A1) | ||
| Meniere's Disease | Syndromic | AD | COCH | Postlingual | COCH5B2 | Brigham and Women's Hospital (7030235), Brigham and Women's Hospital & U-Antwerp (6730475) | ||
| Cockayne Syndrome Type A | Syndromic | ERCC8 | Prelingual | ERCC8 | ||||
| Cockayne Syndrome Type B | Syndromic | ERCC6 | Prelingual | ERCC6 | ||||
| Diabetes-Deafness Syndrome | Syndromic | MTND5, MTTL1 | MTTL1 | |||||
| Charcot-Marie Tooth Neuropathy Type 1A | Syndromic | AD | PMP22 | PMP22 | ||||
| Charcot-Marie Tooth Neuropathy Type 1B | Syndromic | AD | MPZ | MPZ | ||||
| Charcot-Marie Tooth Neuropathy Type 1C | Syndromic | AD | LITAF | LITAF | ||||
| Charcot-Marie Tooth Neuropathy Type 1D | Syndromic | AD | EGR2 | EGR2 | ||||
| Charcot-Marie Tooth Neuropathy Type 1E | Syndromic | AD | PMP22 | PMP22 | Athena (5691144), Athena (6001576) | |||
| Charcot-Marie Tooth Neuropathy Type 1F/2E | Syndromic | AD | NEFL | NEFL | ||||
| Isolated Renal Hypomagnesemia | Syndromic | CLDN16, | CLDN16, | |||||
| Urticaria-Deafness-Amyloidosis (UDA) Syndrome | Syndromic | CIAS1, NLRP3 | CIAS1, NLRP3 | |||||
| Long QT Syndromes and Deafness | Syndromic | KVLQT1, SCN5A | KVLQT1, SCN5A | U-Utah Research Foundation (20020061524A1), U-Utah Research Foundation and Genzyme, Inc (6582913), U-Utah Research Foundation (6787309) | ||||
| Jervell and Lange Nielsen (JLN) Syndrome | Syndromic | AR | KLVQT1, KCNQ1 (JLN1), KCNE1 (JLN2) | Prelingual | Rare | KLVQT1, KCNQ1 (JLN1), KCNE1 (JLN2) | U-Utah Research Foundation (6150104) | |
| Stickler Syndrome | Syndromic | AD | COL11A2, COL2A1, COL11A1, COL9A1 | COL11A1, COL11A2, COL2A1, COL9A1 | ||||
| Epstein Syndrome | Syndromic | AD | MYH9 | MYH9 | ||||
| Norrie Disease | Syndromic | NDP | NDP | |||||
| Treacher Collins Syndrome | Syndromic | TCOF1 | TCOF1 | |||||
| Usher Syndrome Type I (USH1) | Syndromic | AR | MY07A,O USH1C, CDH23, PCDH15, SANS | Prelingual | all Usher combined 3–6% of child deafness | all Usher combined 4.4/100,000 | MY07A, PCDH15 | |
| Usher Syndrome Type II (USH2) | Syndromic | AR | USH2A, VLGR1, WHRN | Prelingual | all Usher combined 3–6% of child deafness | all Usher combined 4.4/100,001 | USH2A | |
| Usher Syndrome Type III (USH3) | Syndromic | AR | USH3 | Postlingual | all Usher combined 3–6% of child deafness | all Usher combined 4.4/100,002 | USH3A (CLRN1) | |
| Kearns-Sayre Syndrome | Syndromic | Mitochondrial | mtDNA deletion syndromes | |||||
| Pearson Syndrome | Syndromic | Mitochondrial | mtDNA deletion syndromes | |||||
| Progressive External Ophthalmoplegia | Syndromic | Mitochondrial | mtDNA deletion syndromes | |||||
| Leigh Syndrome | Syndromic | Mitochondrial | MTATP6, MTTL1, MTTK, MTND1, MTND3, MTND4, MTND5, MTND6, MTCO3, MTTW, and MTTV | MT-ATP6, MT-CO3, MT-ND1, MT-ND3, MT-ND4, MT-ND5, MT-ND6, MT-TK, MT-TL1, MT-TV, MT-TW | ||||
| NARP | Syndromic | Mitochondrial | MTATP6 | MT-ATP6 | ||||
| MELAS | Syndromic | Mitochondrial | MTTL1, MTND5, MT-TC, MT-TV, MT-TF, and MT-TS1 | MTTL1, MTND5 | ||||
| MERRF | Syndromic | Mitochondrial | MTTK | MTTK | ||||
| Vohwinkel Syndrome | Syndromic | GJB2 (Cx 26) | GJB2 >50% | GJB2 (Cx 26) | Institut Pasteur (5998147, 6485908) | |||
| Deafness-Dystonia Syndrome (DDON) | Syndromic | XL | TIMM8A | Varies | ||||
| Hypoparathyroidism, Sensorineural Deafness, and Renal (HDR) Disease | Syndromic | GATA3 | GATA3 | |||||
| Ichthyosis, Hystrix-like, with Deafness | Syndromic | GJB2 (Cx 26) | GJB2 (Cx 26) | Institut Pasteur (5998147, 6485908) |
APPENDIX III
UNIVERSAL NEWBORN HEARING SCREENING EDHI GUIDELINES FOR PEDIATRIC MEDICAL HOME PROVIDERS
See guidelines available at http://www.infanthearing.org/medicalhome/aap_gpmhp.pdf [accessed January 16, 2009].
APPENDIX IV
CINCINNATI CHILDREN’S HOSPITAL HEARING LOSS GENETIC EVALUATION CLINICAL GUIDELINES
[IMAGE]
See http://www.cincinnatichildrens.org/assets/0/78/1067/3345/3399/3403/c2eb6159-2c26-43c6-b2cc-0db5785eb0d5.pdf [accessed January 19, 2009]. Image copyright Cincinnati Children’s Hospital Medical Center, used with permission.
APPENDIX V: PATENT APPLICATIONS FOR HIGH THROUGHPUT HEARING LOSS DIAGNOSTIC TESTING
| Patent/Application No. |
Assignee | Inventors | Publication/File Date |
Title |
|---|---|---|---|---|
| US20070009887A1 | None | Victoria Siemering, Henrik Dahl | 2007-01-11/2003-11-18 | Genotyping of deafness by oligonucleotide microarray analysis |
| US20070134691A1 | None | Iris Schrijver et al.(Stanford Univ, CA) | 2007-06-14/2006-11-14 | Methods & compositions for determining whether a subject carries a gene mutation associated with hearing loss. |
| US20050112598A1 US20040166495A1 | None | Greinwald, John H. Wenstrup, Richard J Aronow, Bruce J. | 2005-05-26 / 2004-02-24 2004-08-26 / 2003-02-24 | Microarray-based diagnosis of pediatric hearing impairment-construction of a deafness gene chip |
| US20040038266A1a | None | Dobrowolski, Steven F Lin, Zhili | 2004-02-26 / 2003-05-22 | Advancing the detection of hearing loss in newborns through parallel genetic analysis |
| US20050059041A1 | None | Johnson, Robert C. Mohammed, Mansoor Kim, Jae Weon Lu, Xan-Yan | 2005-03-17 / 2004-05-17 | Nucleic acids arrays and methods of use therefore |
| US20040203035A1 | Third Wave Technologies, Inc. | Mast, Andrea L. Dorn, Erin; Kwiatkowski, Robert J Accola, Molly Wigdal, Susan S | 2004-10-14 / 2004-01-09 | Connexin allele detection assays |
Inventors Steven F.Dobrowolski and Zhili Lin were employees of NeoGen Screening Inc which was acquired by Pediatrix Medical Group and renamed Pediatrix Screening in 2003.
APPENDIX VI
LETTER FROM GENEDX TO ATHENA DIAGNOSTICS REGARDING CONNEXIN 26 SEQUENCING
October 11, 2006
Michael W. Henry
VP, Business Development
Athena Diagnostics, Inc.
Four Biotech Park
377 Plantation Street
Worcester, MA 01605
Re: GeneDx testing in GJB2 gene
Dear Mr. Henry:
I am in receipt of your letter dated September 11, 2006 regarding Athena Diagnostics being the exclusive licensee of two US Patents. You noted that a letter had been sent to John Compton on November 11, 2002 regarding this issue. That letter was never received at GeneDx or by John Compton. We had moved to 207 Perry Parkway the previous month and our mail was not being forwarded by the post office.
We have reviewed the two patents to which your letter of September 11, 2006 refer (5,998,147 and 6,485,908). These two patents specifically make claims regarding detecting mutations in the Connexin 26 gene comprising a deletion of a nucleotide from nucleotides 30 to 32 or a deletion of 38 base pairs beginning at position 30 (mutations described as being involved in autosomal recessive prelingual non-syndromic deafness).
GeneDx provides GJB2 (Connexin 26) gene testing for the ectodermal dysplasia known as Keratitis-Ichthyosis-Deafness syndrome, a severe and sometimes lethal autosomal dominant syndromic disorder. KID syndrome is considered an ultra-rare disorder, with only about 100 cases reported in the literature. Nearly 80% of patients with KID syndrome have a mutation, D50N, in the GJB2 gene. The mutation spectrum in KID syndrome and other rare dominant syndromic disorders involving the GJB2 gene have been published by the principals of GeneDx (see below).
You can find the details of the testing we offer on our website (www.genedx.com) and in the information sheet that can be downloaded from the site. Should you have any further questions, please do not hesitate to contact us.
Sincerely,
Sherri J. Bale, Ph.D., FACMG
Clinical Director
GeneDx
Richard G, White TW, Smith LE, Bailey RA, Compton JG, Paul DL, Bale SJ. Functional defects of Cx26 resulting from a heterozygous missense mutation in a family with dominant deaf-mutism and palmoplantar keratoderma. Hum Genet. 1998 Oct;103(4):393-9.
Richard G, Rouan F, Willoughby CE, Brown N, Chung P, Ryynanen M, Jabs EW, Bale SJ, DiGiovanna JJ, Uitto J, Russell L. Missense mutations in GJB2 encoding connexin-26 cause the ectodermal dysplasia keratitis-ichthyosis-deafness syndrome.
Am J Hum Genet. 2002 May;70(5):1341-8. Epub 2002 Mar 22.
Richard G, Brown N, Ishida-Yamamoto A, Krol A. Expanding the phenotypic spectrum of Cx26 disorders: Bart-Pumphrey syndrome is caused by a novel missense mutation in GJB2. J Invest Dermatol. 2004 Nov;123(5):856-63.
APPENDIX VII
EMAIL RESPONSE FROM ATHENA DIAGNOSTICS TO GENE DX SHARED WITH PERMISSION OF DR. SHERRI BALE, CLINICAL DIRECTOR, GENEDX
Delivered-To: sherrib@genedx.com
Tue, 31 Oct 2006 15:31:28 -0500
Dear Sherri:
Thank you for your attached letter of October 11 regarding Cx26 (GJB2).
I understand that you test for the GJB2 D50N mutation for Keratitis-Icthyosis-Deafness (KID) syndrome.
Please confirm that GeneDx GJB2 testing does not include testing for the following mutations:
Deletion of nucleotides 27–35
Deletion of 38 base pairs starting at position 30
Deletion at position 30
Deletion of a nucleotide from nucleotide 30 to nucleotide 32
If GeneDx does not test for these GJB2 mutations, then I will consider this matter closed.
Regards,
Mike
Michael W. Henry
Vice President, Business Development
Athena Diagnostics, Inc.
377 Plantation Street
Worcester, MA 01605
(508) 756-2886 x3100
(508) 752-7421 fax
mhenry@athenadiagnostics.com
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
HUMAN STUDIES: All interviews were conducted under Duke University IRB-approved protocol 1277 and usually conducted by phone and recorded. Researchers obtained informed consent from subjects.
Contributor Information
Subhashini Chandrasekharan, Center for Genome Ethics, Law & Policy, Institute for Genome Sciences & Policy, Duke University.
Melissa Fiffer, Center for Genome Ethics, Law & Policy, Institute for Genome Sciences & Policy and Nicholas School of the Environment and Earth Sciences, Duke University.
References
- 1.Burton S, Withrow K, Arnos K, Kalfoglou A, Pandya A. A focus group study of consumer attitudes toward genetic testing and newborn screening for deafness. Genet Med. 2006;8(12):779–783. doi: 10.1097/01.gim.0000250501.59830.ff. [DOI] [PubMed] [Google Scholar]
- 2.Taneja P, Pandya A, Foley D, Nicely L, Arnos K. Attitudes of deaf individuals towards genetic testing. Am J Med Genet. 2004;130(1):17–21. doi: 10.1002/ajmg.a.30051. [DOI] [PubMed] [Google Scholar]
- 3.American College of Medical Genetics. Genetics evaluation guidelines for the etiologic diagnosis of congenital hearing loss. Genet Med. 2002;4(3):162–171. doi: 10.1097/00125817-200205000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heller M, Eisenberg R. Can patents deter innovation? The anticommons in biomedical research. Science. 1998 May 1;280(5364):698–701. doi: 10.1126/science.280.5364.698. [DOI] [PubMed] [Google Scholar]
- 5.Transcript of a conference call held by Hologic, Inc on June 9, 2008. SEC Accession No. 0001193125-08-130599; SEC.2008. [Google Scholar]
- 6.Boston University and The Center for Human Genetics, Inc. Announce Change in Testing Services. [Accessed November 14, 2008];BioMedicine News. August 18; http://www.bio-medicine.org/medicine-technology-1/Boston-University-and-The-Center-for-Human-Genetics--Inc--Announce-Change-in-Testing-Services-2851-1/
- 7.Morton C, Nance W. Newborn hearing screening - a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 8.Kelsell D, Dunlop J, Stevens H, et al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature. 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- 9.Denoyelle F, Weil D, Maw M, et al. Prelingual deafness: high prevalence of 30delG mutation in the connexin 26 gene. Hum Mol Genet. 1997;6:2173–2177. doi: 10.1093/hmg/6.12.2173. [DOI] [PubMed] [Google Scholar]
- 10.Nanogen licenses rights to gene linked to hereditary deafness. [Accessed March 10, 2007];Fresh News. 2003 http://www.freshnews.com/news/biotech-biomedical/article_15724.html?Nanogen.
- 11.Institut Pasteur Technology Transfer. [Accessed January 16, 2009]; http://www.pasteur.fr/ip/easysite/go/03b-000024-0ap/licensing-opportunities/list-oftechnologies-available-for-licencing-or-transfer.
- 12.Kenneson A, Myers M, Lubin I, Boyle C. Genetic laboratory practices related to testing of the GJB2 (connexin 26) gene in the United States in 1999 and 2000. Genet Test. 2003;7(1):49–56. doi: 10.1089/109065703321560949. [DOI] [PubMed] [Google Scholar]
- 13.Pandya A, Arnos K, Xia X, et al. Frequency and distribution of GJB2 (connexin 26) and GJB6 (connexin 30) mutations in a large North American repository of deaf probands. Genet Med. 2003;5(4):295–303. doi: 10.1097/01.GIM.0000078026.01140.68. [DOI] [PubMed] [Google Scholar]
- 14.University of Cardiff. Human gene mutation database. [Accessed November 19, 2008]; http://www.hgmd.cf.ac.uk/ac/gene.php?gene=GJB2.
- 15.Castillo Id, Villamar M, Moreno-Pelayo M, et al. A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N Engl J Med. 2002;346(243–249):243–249. doi: 10.1056/NEJMoa012052. [DOI] [PubMed] [Google Scholar]
- 16.Putcha G, Bejjani B, Bleoo S, et al. A multicenter study for the frequency and distribution of GJB2 and GJB6 mutations in a large North American cohort. Genet Med. 2007;9(7):413–426. doi: 10.1097GIM.0b013e3180a03276. [DOI] [PubMed] [Google Scholar]
- 17.Everett L, Glaser B, Beck J, et al. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS) Nat Genet. 1997;17:411–422. doi: 10.1038/ng1297-411. [DOI] [PubMed] [Google Scholar]
- 18.Smith R, van Camp G. Pendred syndrome/DFNB4. [Accessed April 15, 2007];GeneReviews. http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=pendred.
- 19.Cook-Deegan RM, DeRienzo C, Carbone J, Chandrasekharan S, Heaney C, Conover C. Impact of gene patents on access to genetic testing for inherited susceptibility to cancer: comparing breast and ovarian cancers to colon cancers. Genet Med. 2010 doi: 10.1097/GIM.0b013e3181d5a67b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandya A. Nonsyndromic hearing loss and deafness, mitochondrial. [Accessed January 15, 2008];GeneReviews. http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=mt-deafness.
- 21.Prezant T, Agapian J, Bohlman M, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 22.Estivill X, Govea N, Barcelo A, et al. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment with aminoglycosides. Am J Hum Genet. 1998;62:27–35. doi: 10.1086/301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid F, Vernham G, Jacobs H. A novel mitochondrial point mutation in a maternal pedigree with sensorineural deafness. Hum Mutat. 1994;3(3):243–247. doi: 10.1002/humu.1380030311. [DOI] [PubMed] [Google Scholar]
- 24.National Institutes of Health; 1993. Mar 1–3, [Accessed January 16, 2009]. Early identification of hearing impairment in infants and young children: National Institutes of Health Consensus Development Conference Statement. http://consensus.nih.gov/1993/1993HearingInfantsChildren092html.htm. [Google Scholar]
- 25.White K. The current status of EHDI programs in the United States. Ment Retard Disabil Res Rev. 2003;9:79–88. doi: 10.1002/mrdd.10063. [DOI] [PubMed] [Google Scholar]
- 26.National Resource Center for Early Hearing Detection and Intervention. [Accessed August, 2007]; http://www.infanthearing.org/tas/index.html.
- 27.Norris V, Arnos K, Hanks W, et al. Does universal newborn hearing screening identify all children with GJB2 (connexin 26) deafness? Penetrance of GJB2 deafness. Ear & Hearing. 2006;27:732–741. doi: 10.1097/01.aud.0000240492.78561.d3. [DOI] [PubMed] [Google Scholar]
- 28.Arnos K. The implications of genetic testing for deafness. Ear & Hearing. 2003;24:324–331. doi: 10.1097/01.AUD.0000079800.64741.CF. [DOI] [PubMed] [Google Scholar]
- 29.White K. Early hearing detection and intervention programs: opportunities for genetic services. Am J Med Genet. 2004;130A(1):29–36. doi: 10.1002/ajmg.a.30048. [DOI] [PubMed] [Google Scholar]
- 30.Schimmenti L, Martinez A, Fox M. Genetic testing as part of the early hearing detection and intervention (EHDI) process. Genet Med. 2004;6(6):521–525. doi: 10.1097/01.gim.0000144187.21727.28. [DOI] [PubMed] [Google Scholar]
- 31.Robin N, Prucka S, Woolley A, Smith R. The use of genetic testing in the evaluation of hearing impairment in a child. Curr Opin Pediatr. 2005;17:709–712. doi: 10.1097/01.mop.0000181469.56484.5f. [DOI] [PubMed] [Google Scholar]
- 32.Connor C, Craig H, Raudenbush S, Heavner K, Zwolan T. The age at which young deaf children receive cochlear implants and their vocabulary and speech-production growth: is there an added value for early implantation? Ear Hear. 2006;27(6):628–644. doi: 10.1097/01.aud.0000240640.59205.42. [DOI] [PubMed] [Google Scholar]
- 33.A white paper addressing the societal costs of hearing loss and issues in third party reimbursement. [Accessed January 16, 2009];2004 November 8; http://www.audiologyonline.com/articles/pf_article_detail.asp?article_id=1204.
- 34.Mohr P, Feldman J, Dunbar J, et al. The societal costs of severe to profound hearing loss in the United States. Int J Technol Assess Health Care. 2000;16(4):1120–1135. doi: 10.1017/s0266462300103162. [DOI] [PubMed] [Google Scholar]
- 35.Preciado D, Lim L, Cohen A, et al. A diagnostic paradigm for childhood idiopathic hearing loss. Otolaryngol Head Neck Surg. 2004;131:804–809. doi: 10.1016/j.otohns.2004.06.707. [DOI] [PubMed] [Google Scholar]
- 36.Fischel-Ghodsian N. Mitochondrial mutations and hearing loss: paradigm for mitochondrial genetics. Am J Hum Genet. 1998;62:15–19. doi: 10.1086/301695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joint Committee on Infant Hearing (JCIH) Year 2000 position statement: Principles and guidelines for early hearing detection and intervention programs. Pediatrics. 2000;106(4):798–817. doi: 10.1542/peds.106.4.798. [DOI] [PubMed] [Google Scholar]
- 38.Pediatrix Medical Group. SoundGene Clinician Information. [Accessed January 16, 2009]; http://www.pediatrix.com/body.cfm?id=2889.
- 39.Siemering K, Manji S, Hutchison W, Du Sart D, Phelan D, Dahl H. Detection of mutations in genes associated with hearing loss using a microarray-based approach. J Mol Diagn. 2006;8(4):483–489. doi: 10.2353/jmoldx.2006.050147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardner P, Oitmaa E, Messner A, Hoefsloot L, Metspalu A, Schrijver I. Simultaneous multigene mutation detection in patients with sensorineural hearing loss through a novel diagnostic microarray: A new approach for newborn screening follow-up. Pediatrics. 2006;118(3):985–994. doi: 10.1542/peds.2005-2519. [DOI] [PubMed] [Google Scholar]
- 41.Shumaker J, Metspalu A, Caskey C. Mutation detection by solid phase primer extension. Hum Mutat. 1996;7(4):346–354. doi: 10.1002/(SICI)1098-1004(1996)7:4<346::AID-HUMU9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 42.Kurg A, Tonisson N, Georgiou ISJ, Tollett J, Metspalu A. Arrayed primer extension: Solid-phase four-color DNA resequencing and mutation detection technology. Genet Test. 2000;4(1):1–7. doi: 10.1089/109065700316408. [DOI] [PubMed] [Google Scholar]
- 43.Asper Biotech. Hereditary Hearing Loss Testing. [Accessed March 10, 2007]; http://www.asperbio.com/HHL.htm.
- 44.Asper Biotech. Hereditary Hearing Loss Tests Supplement. [Accessed March 10, 2007]; www.asperbio.com/supplement.pdf.
- 45.Preciado D, Lawson L, Madden C, et al. Improved diagnostic effectiveness with a sequential diagnostic paradigm in idiopathic pediatric sensorineural hearing loss. Otol Neurotol. 2005;26(4):610–615. doi: 10.1097/01.mao.0000178133.89353.1d. [DOI] [PubMed] [Google Scholar]
- 46.Computational Medical Center. Gene chip diagnostic test for pediatric hearing impairment. [Accessed March 10, 2007]; http://www.computationalmedicine.org/project/hearing_loss.htm.
- 47.Cai-Xia I, Qian P, Guo Y-G, et al. Construction of a multiplex allele-specific PCR-based universal array (ASPUA) and its application to hearing loss screening. Hum Mutat. 2008;29:306–314. doi: 10.1002/humu.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.House Judiciary Subcommittee on Courts, the Internet and Intellectual Property. Washington, DC: 2007. [Accessed February 4, 2010]. Statement of Marc Grodman at Stifling or Stimulating – The Role of Gene Patents in Research and Genetic Testing. http://judiciary.house.gov/hearings/pdf/Grodman071030.pdf. [Google Scholar]
- 49.Moeller M, White K, Shisler L. Primary care physicians' knowledge, attitudes, and practices related to newborn hearing screening. Pediatrics. 2006;118(4):1357–1370. doi: 10.1542/peds.2006-1008. [DOI] [PubMed] [Google Scholar]
- 50.Duncan R, Prucka S, Wiatrak B, Smith R, Robin N. Pediatric otolaryngologists' use of genetic testing. Arch Otolaryngol Head Neck Surg. 2007;133(3):231–236. doi: 10.1001/archotol.133.3.231. [DOI] [PubMed] [Google Scholar]
- 51.CIGNA HealthCare. Cigna medical coverage policy. [Accessed January 16, 2009]; http://www.cigna.com/customer_care/healthcare_professional/coverage_positions/medical/mm_0254_coveragepositioncriteria_genetic_test_congenital_profound_deafness.pdf.
- 52.Athena Diagnostics. Ordering & billing. [Accessed August, 2007]; http://www.athenadiagnostics.com/content/ordering/
- 53.Castillo Id, Moreno-Pelayo MA, del Castillo FJ, et al. Prevalence and evolutionary origins of the del(GJB6-D13S1830) mutation in the DFNB1 locus in hearing-impaired subjects: a multicenter study. Am J Hum Genet. 2003;73(6):1452–1458. doi: 10.1086/380205. [DOI] [PMC free article] [PubMed] [Google Scholar]