Fig. 1.
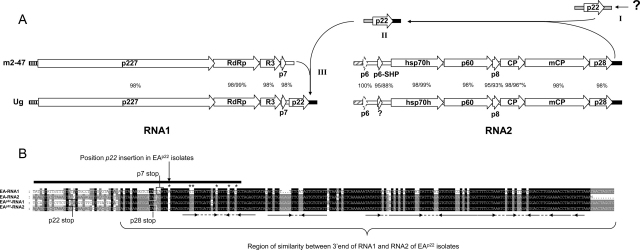
(A) Schematic representation and comparison of Sweet potato chlorotic stunt virus genome organization of isolates m2-47 and Ug. Percentage values represent the identity at nucleotide level for each ORF; proteins identities are given behind the slash if different from nucleotide. * includes 3nt/1aa deletion in the CP of isolate m2-47. Different patterns of shading at the terminal parts of the genomes of both isolates represent differences in sequence conservation at these regions. There is a second ORF encoding a putative protein with transmembrane domain (p6-SHP) in m2-47 and the six other isolates (Table 1) sequenced in this region, the percentages shown correspond to its comparison with the same region in isolate Ug, except that a single nucleotide change (A575 → T) interrupts the p6-SHP in Ug (indicated by a question mark). The p6 is absent from isolates (Africa10 and Bitambi) and is indicated by dotted lines. Arrows show a suggested route for p22 acquisition, with roman numerals indicating a suggested sequence of steps giving origin to p22 in isolate Ug. A question mark indicates the unknown source of p22. (B) Alignment of the 3′-UTRs of EA and EAp22 isolates characterized in this study (Table 1) highlighting the dissimilarity in this region. The bold line above the alignment on the left indicates the region of RNA1-3′-UTR sequenced previously for other isolates lacking p22 (Cuellar et al., 2008), and nucleotides conserved between those and m2-47 are indicated with an asterisk. Arrows below the alignment indicate conserved secondary structure of the 3′-UTR found in all criniviruses; stop codons of the 3′ terminal ORFs are indicated by boxes. The last 10 nts of m2-47 RNA2 were not sequenced.