Abstract
A comprehensive model of evolution requires an understanding of the relationship between selection at the molecular and phenotypic level. We investigate this in Strepsiptera, an order of endoparasitic insects whose evolutionary biology is poorly studied. We present the first molecular phylogeny of Strepsiptera, and use this as a framework to investigate the association between parasitism and molecular evolution. We find evidence of a significant burst in the rate of molecular evolution in the early history of Strepsiptera. The evolution of morphological traits linked to parasitism is significantly correlated with the pattern in molecular rate. The correlated burst in genotypic-phenotypic evolution precedes the main phase of strepsipteran diversification, which is characterised by the return to a low and even molecular rate, and a period of relative morphological stability. These findings suggest that the transition to endoparasitism led to relaxation of selective constraint in the strepsipteran genome. Our results indicate that a parasitic lifestyle can affect the rate of molecular evolution, although other causal life-history traits correlated with parasitism may also play an important role.
Introduction
A central focus in evolutionary research is the interaction between molecular evolution and selection at the level of the phenotype, the interface of which unifies aspects of evolutionary research often examined independently [1]. Such an approach offers insight into the factors shaping the rate of molecular evolution, and into the link between genome evolution and species divergence [2]. Strepsiptera is an order of insect parasitoids which display a variety of unusual genetic and phenotypic features [3]–[8]. Targeting groups with complex biologies such as Strepsiptera is useful for testing the validity and generality of ecological and evolutionary theory [9], [10]. However, insufficient molecular data have prevented the study of a number of interesting questions, such as the relationship between genotypic and phenotypic evolution. Strepsiptera display characteristics that are close to the parasite/parasitoid boundary [4], [11]. Female morphology is highly derived (eyes, antennae, mouthparts, legs, wings and reproductive characters are lost) and accompanied by an endoparasitic lifestyle that is host-dependent throughout the lifecycle (except for the family Mengenillidae). In contrast the male is free-flying as an adult and possesses typical insect characteristics. Strepsiptera infect a broad range of hosts, and are recorded from at least 34 families of insects distributed across 7 orders [3], [4], [12]. As with many other parasitic taxa, relatively little research has examined the evolution of host-usage in the group and its effect on speciation. However, recognition of the contribution of parasitic taxa to total animal diversity [13] has emphasised the need to understand the basis of parasite diversification and host usage [14].
The relationship between genotype and phenotype can be examined in a variety of ways. Positive selection in candidate gene phylogenies has been paired with extant phenotypic traits on terminal or internal branches of a phylogeny [15], [16]. Alternatively, a null model of evolution can be compared against models that specify positive selection [17]. These methods specifically target associations in genes responsible for particular phenotypic adaptations (i.e. those under positive selection). Another approach focuses instead on the differing molecular evolutionary rates between species, and in identifying potential life-history traits that influence molecular evolution. Understanding is limited by the availability of suitable methodology, since the field has emerged recently in response to increased DNA sequence data [2]. Investigations have searched for meaningful associations between the rate of molecular evolution and key phenotypic or other extrinsic factors [18], [19], or between phenotypic factors and significant shifts in the pattern of lineage diversification [20]. Results from such studies must be carefully interpreted [21], [22], due to errors associated with phylogeny and rate estimation, or ancestral state reconstruction [1], [2], [23].
Here we investigate the nature and underlying cause of a common feature of higher-level insect phylogenetic analyses: the long-branch separating Strepsiptera from other insect groups [24]–[30]. We explore the link between the strepsipteran phenotype's evolution and: i) variation in the rate of molecular evolution: ii) the pattern of lineage diversification. We reconstruct the first robust molecular phylogeny of Strepsiptera, and use this as a framework to investigate the history of morphological and host-use evolution. Key characteristics include the loss of compound eyes, antennae, legs, wings and reproductive structures in the female, and modifications to the legs and tarsi, and loss of mandibles in some males. The questions we address include the point at which strepsipteran traits evolved, if they emerged more than once and how they are associated with variation in molecular evolution. We explore the hypothesis that the parasitic lifestyle exerts an effect on the rate of molecular evolution [31], an assertion that few studies have so far been able to support [32], [33]. To do so, we examine molecular rate variation in Strepsiptera, and establish a model of evolutionary history that encompasses both molecular and phenotypic evolution.
Results
Strepsiptera phylogeny
We used 41 strepsipteran taxa, across 16 genera, and data from four genes: the mitochondrial genes cytochrome c oxidase I (cox1), NADH dehydrogenase I (nad1), and small subunit ribosomal RNA (16S rRNA), and the nuclear gene small subunit ribosomal RNA (18S rRNA) to generate a final alignment of 3930 nucleotides. This consisted of 967 bp 803 bp, and 2160 bp for the 18S rRNA, 16S rRNA, and cox1+nad1 partitions respectively.
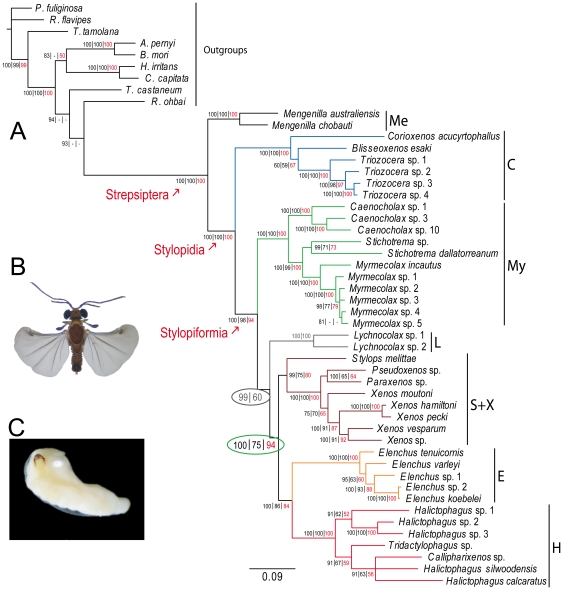
Across all phylogenetic analyses, we identified monophyletic groupings for extant Strepsiptera, Stylopidia and Stylopiformia, corroborating the findings from previous studies [3], [4], [12], [34] using molecular data for the first time. In the concatenated Bayesian Inference (BI) analyses, all nodes (bar one) at and above the family level receive 100 posterior support (Figure 1A). Maximum Likelihood (ML) bootstrap values are ≥75/100 with Lychnocolax (except one: node within grey oval); and ≥80/100 without Lychnocolax. We find Myrmecolacidae as the sister-clade to all remaining families within Stylopiformia, and a sister-group relationship between Stylopidae (which parasitize bees) and Xenidae (which parasitize crabronid, sphecid, eumenid and vespid wasps), and between Halictophagidae and Elenchidae that predominantly parasitize Auchenorrhyncha (a “true bug” group, containing amongst others the cicadas, leafhoppers, treehoppers, planthoppers and spittlebugs). The genus Lychnocolax has no host records, and was historically placed within Myrmecolacidae [3]. Here, we find evidence that it is an older taxon, as the sister-group to Stylopidae, Xenidae, Elenchidae and Halictophagidae. Its position in the analyses is supported by ribosomal nucleotide composition data (Figure S1), but this is only moderately supported in the concatenated phylogenetic analyses (Figure 1A, grey oval). Removal of Lychnocolax led to increased ML bootstrap support in a descendent node (Figure 1A; green oval). Genera are all returned as monophyletic except Halictophagus, which occurred as a poorly resolved polyphyletic grouping with Tridactylophagus and Callipharixenos. The latter species is placed within Halictophagidae, arguing against the separate family-status hypothesized for this lineage.
Figure 1. Molecular phylogeny of Strepsiptera.
(A) Branch lengths from the BI 50% majority rule tree, with support values from BI and ML analyses appearing next to nodes. Support values (%) = BI posterior support | 1000 ML parametric bootstraps with Lychnocolax | 500 ML parametric bootstraps without Lychnocolax. Grey oval: support values = BI posterior support | 1000 ML parametric bootstraps including Lychnocolax. Green oval: increased ML support following removal of Lychnocolax. Me = Mengenillidae; C = Corioxenidae; My = Myrmecolacidae; L = Lychnocolax; S+X = Stylopidae+Xenidae; E = Elenchidae; H = Halictophagidae. (B) Male Caenocholax fenyesi sensu lato (C) Female Caenocholax fenyesi sensu lato [4].
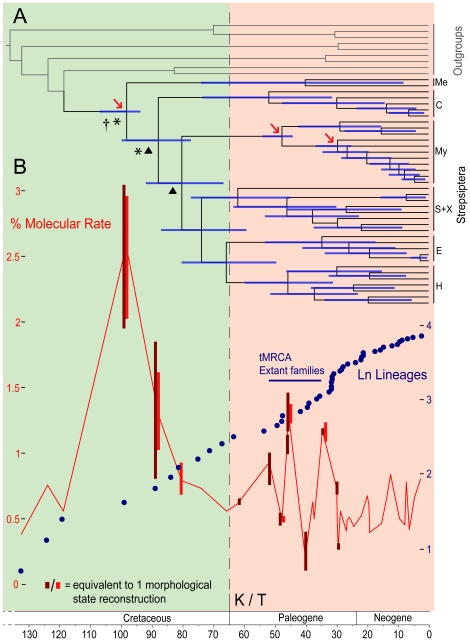
The dated phylogeny based on the MIT1+2 dataset (see methods) is given in Figure 2, with 95% credibility interval (CI) bars positioned over relevant nodes. The tree reflects the topology produced using the entire (concatenated) dataset presented in Figure 1 (based on the nuclear 18S rRNA+ mitochondrial 16S rRNA+ cox1/nad1 partitions), although there is minor incongruence within Myrmecolacidae, Xenidae, Halictophagidae, Elenchidae. Differences between MIT1+2 and MIT123 (the mitochondrial dataset including 3rd codon positions) on date estimation was minor, with a marginal increase in 95% CIs using MIT1+2 (Table S1). Imprecise CIs concentrate in regions less well informed by available fossil prior information.
Figure 2. History of divergence and rate of molecular evolution in Strepsiptera.
(A) BI phylogeny using the MIT1+2 dataset calibrated against time. Node age 95% credibility intervals are indicated over nodes. †Increased relative rate of 18S rRNA. *Increased relative rate of MIT1+2. ▴Significant Relative Cladogenesis (RCT) statistics. Arrows indicate fossil calibrated nodes. Clade abbreviations follow Figure 1. (B) History of molecular rate using MIT1+2 scaled to the tree in panel A with number of ancestral character reconstructions at corresponding nodes (dark red bars = total non-homoplastic state changes in morphology under parsimony [34], red bars = total morphological reconstructions using Bayesian ancestral reconstruction). Blue: Log number of lineages at corresponding distance from root. tMRCA = time to Most Recent Common Ancestor; K/T = Cretaceous/Tertiary boundary.
Molecular evolution
Firstly, we investigated the history of molecular rate across the MIT1+2 and MIT123 phylogenies. We found molecular rate estimates ranging from 1–1.5% pairwise sequence divergence per million years (Table S1) for analyses including 3rd codon positions. These are lower than the commonly cited value of 2.3% [35] and more in-keeping with 1.5% [36] for insect mitochondrial DNA and other rates reported for Strepsiptera [10]. However, within this overall pattern, the dated trees and relative rate analyses revealed significant variation in molecular rate, notably at the time to most recent common ancestor (tMRCA) of Strepsiptera and Stylopidia (Figure 2). Both nodes are associated with clades with high relative rates of molecular evolution. The rates at descendent strepsipteran nodes are lower, at around 1% pairwise sequence divergence per million years. The pattern of molecular evolution in the nuclear 18S rRNA gene is in good overall agreement with the mitochondrial cox1+nad1 gene (MIT1+2, MIT123) datasets (Table S2, Figure S2).
We then compared the evolution of molecular rate with an investigation into the history of diversification rate. The relative cladogenesis test (RCT) indicated a significant shift in Stylopidia and Stylopiformia (Figure 2). But statistics from the topological method in Symmetree were not significant, with upper and lower bound confidence intervals (CI) (at .025 and .975 frequentiles) of 0.079–0.168 and 0.042–0.095 in the MR and MΣ tests respectively. Inclusion of 560 missing taxa produced p-value CIs for whole-tree test statistics (MR; IC; MΠ *; MΠ; MΣ *; MΣ; B1) between 0.001 - 0.000. This discrepancy could stem from over-representation of Mengenillidae and Corioxenidae, and under-representation of Stylopidae, Halictopaghidae and Myrmecolacidae in the taxon set. In both analyses, individual nodes were not associated with rate shifts, with p-values of 0.121 and 0.209 and 0.107 and 0.185 (with missing taxa) for Stylopidia and Stylopiformia respectively. Furthermore, the branching pattern from the maximum clade credibility (MCC) tree did not depart significantly from a constant-rate/null speciation model: 0.999 (b = 0.5, d = 0.5, m = 560); 0.998 (b = 0.5, d = 0.0, m = 560); 0.836 (b = 0.5, d = 0.5, m = 60); 0.701 (b = 0.5, d = 0.0, m = 60). These results indicate that a burst of molecular rate evolution characterised the early evolution of Strepsiptera, but this did not coincide with a significant shift in lineage diversification.
Reconstruction of the strepsipteran phenotype
Having established a basic framework for genotypic evolution, we directed attention towards understanding evolution of the strepsipteran phenotype. The morphological character reconstructions used in BI approach are summarized over the MIT1+2 phylogeny in Figure 2, details of the character state reconstructions that were recovered at each node can be found in Table S3. These corroborate the reconstruction of morphological evolution from a previous phylogeny using parsimony [34]. Both closely mirror the pattern of molecular rate depicted in Figure 2.
The long-branch leading to Strepsiptera is linked with phenotypic modifications relating to extreme sexual dimorphism, obligate endoparasitism in the larval stages, and entomophagy (consumption of insects as food). Stylopidia is associated with the evolution of the endoparasitic female (and the continuation of endoparasitism through pupation for males). In males, this node is linked to the reduction or loss of spiracles in the adult and larvae respectively, and the loss of pupal claws. Stylopiformia is associated with modifications to the tarsi, reduction of tarsal number and loss of larval legs in males, and the evolution of the cephalothorax in females.
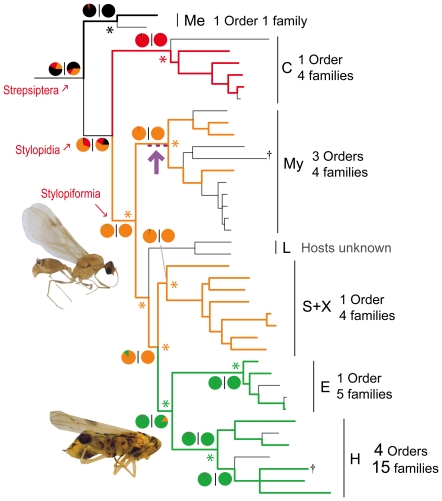
The history of strepsipteran host-use is summarized in Figure 3. Parasitization of aculeate hymenopterans is predicted to have originated in Stylopiformia, or possibly earlier in the ancestor to Stylopidia, where a secondary switch would be implicated in Corioxenidae to Heteroptera (a “true bug” group, containing amongst others the assassin bugs, bed bugs, seed bugs and shield bugs). In both models, a subsequent switch to Auchenorrhyncha in the ancestor of Elenchidae and Halictophagidae is strongly supported. Outside of Myrmecolacidae, host switching between infraordinal host groups occurs only in Halictophagidae. The ancestral host of Strepsiptera remains unresolved given currently available data.
Figure 3. Bayesian reconstruction of male host-usage according to infra-ordinal grouping.
Known host records are given next to clades. Unshaded lines = unknown records/equivocal reconstructions. Black = Lepismatidae; Red = Heteroptera; Orange = Hymenoptera; Green = Auchenorrhyncha; Purple = possible origin of heteronomy. *Significant node reconstructions using BFs. Pie charts = posterior probability | ML support. BFs and support charts not shown below family. †Probable parthenogens. Clade abbreviations follow Figure 1B. Images: Pheidole sp. (Hymenoptera) with male cephalotheca (top). Sogatella furcifera (Homoptera) with Elenchus japonicas male puparium. Photographs © J. Kathirithamby.
Comparison of molecular and phenotypic rates of evolution
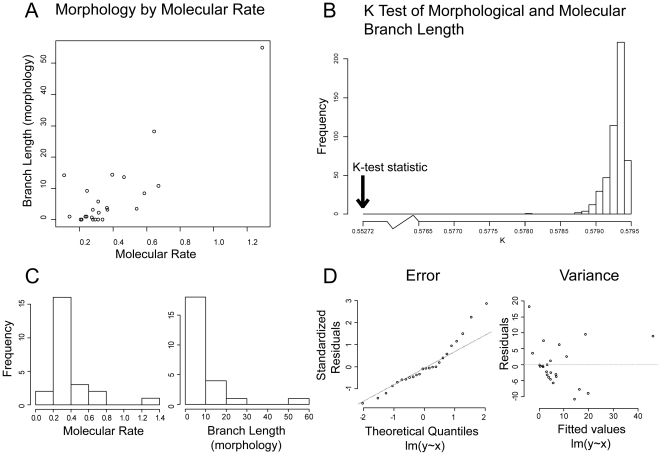
We undertook a number of analyses to test the statistical validity of the association between molecular and phenotypic rates of evolution. A linear model indicated that molecular rate is positively associated with morphological branch length variation (T-statistic = 7.360, p-value = 2.29E-07). Molecular rate contributed the majority of variation in branch length (Adjusted R-squared = 0.698). However, non-linearity of error and heterogeneity of variance undermined the assumptions of a parametric statistical approach. The concentration of molecular and morphological rate evolution in the node leading to Strepsiptera represents a significant component of the skew in the distribution (Figure 4). We therefore re-examined the correlation by using a spearman test (rho = 0.542, S = 1054.628, p-value = 0.003) (Figure 4A).
Figure 4. Graphical summary of molecular rate and morphological branch length variation.
(A) Individual node comparison of molecular rate (pairwise sequence divergence / million years) versus morphological branch length (steps required under parsimony). (B) K-tree null distribution, with test-statistic indicated by arrow. (C) Distribution of molecular rate (left) and morphological branch length (right) variation. (D) Graphical summary of linear model assumptions: non-linearity of error (left) and heterogeneity of variance (right).
We also took an alternative approach by making a whole-tree comparison of morphological versus molecular branch lengths (instead of molecular rate). We generated a null distribution of K-scores (mean = 0.579, S.D = 0.000134, min = 0.5780, max = 0.5794), and compared our K-test statistic of 0.553 against this distribution (see methods). The hypothesis that the observed K-score was due to random processes could be rejected (p-value≪0.001) (Figure 4B). Overall, these results indicate a correlated pattern of molecular and morphological evolution in Strepsiptera.
Discussion
In this study, we develop a framework for understanding strepsipteran molecular evolutionary history, link it with existing knowledge of strepsipteran morphological evolution, and establish a foundation for further research into the evolution and ecology of this unusual host-parasite system. We retrieve high support for the monophyly of Strepsiptera, Stylopidia and Stylopiformia, and for interrelationships between the extant families. There remain areas of uncertainty, in particular the equivocal position of Lychnocolax and the polyphyly of Halictophagus, which require taxonomic revision through re-analysis of morphology and inclusion of additional taxa and alternative DNA markers.
Molecular rate, diversification rate, and phenotypic evolution
We detected a significant shift in molecular rate in the early history of Strepsiptera. Instead of remaining uniformly high, the rate returned to a low and even rate across the phylogeny. Minor peaks in molecular rate are also linked with the diversification of lineages post K/T, in particular the tMRCAs of extant families Xenidae, Halictophagidae, and Elenchidae between 50-30 MYA. Interestingly, the K/T boundary was not closely linked with shifts in either molecular rate or diversification rate. A similar pattern has also been observed in other terrestrial animal groups, including mammals [37], squamates and passerine birds [38]. This trend reflects contemporaneous changes in strepsipteran morphology, which also changed significantly during the group's early history, followed by a period of stability in more recent history (<70 MYA), notwithstanding minor modifications to morphology linked with the tMRCAs of several extant families (Figure 2). Relationships within Stylopiformia are inconsistent with the only other (morphology-based) phylogenetic analysis of Strepsiptera. This important result may be due to the low number of non-homoplasious morphological state changes at intermediate depths of the prior study (Figure 29 in [34]). In the current study, inter-node distances are short compared with surrounding branches in the equivalent region of the tree (Figure 1). This is consistent with the individual node comparison of molecular rate and morphological branch length, which show a positive correlation.
The evolution of strepsipteran structural morphology involved a range of adaptations associated with increasingly specialised parasitism. Extant Mengenillidae represent a transitional condition, in that females reproduce and release progeny whilst outside of the host, with the faculty to leave the free-living pupa to lead a motile lifestyle [39]. We hypothesize that complete female endoparasitism in Stylopidia led to strong sexual selection on free-living adult males, which to copulate successfully must engage with highly modified female structures (the cephalothorax) protruding from living hosts. This may have led to the evolution of hairy adhesive tarsal pads; needed to adhere to diverse host substrates during insemination of the endoparasitic female [40].
The diversification rate in Strepsiptera increased after the initial burst in molecular rate, but a significant individual node shift was only detected in the RCT statistic, at the origin of Stylopidia/Stylopiformia. Lack of evidence for an increase in diversification across other methods suggests this result should be interpreted with caution. We refrain from discussing in depth which phenotypic traits (if any) might be causally linked to the main phase of strepsipteran diversification due to difficulties associated with identifying trait(s) that are responsible for speciation [41], [42]. One might hypothesize that after endoparasitism became an obligate component of all aspects of female life-history (in the ancestor of Stylopidia), the evolution of a more effective method of host immune evasion may have enhanced the ability of Strepsiptera to successfully infect novel hosts, thereby opening opportunities for speciation. During infection, Strepsiptera are contained within a host-derived epithelial membrane, which is thought to conceal the endoparasite from the host's immune system [5]. However, its point of origin remains unknown.
Did parasitism cause the burst in molecular rate?
We discovered a correlated burst in the rate of molecular and morphological evolution, which coincided with significant increases in the relative rate of molecular evolution, and abrupt shifts in the evolution of rRNA structures (Figure S1). We showed in an overall comparison of branch length that the observed similarity between molecular and morphological trees (K-score) was not due to random processes. The traits that evolved during the early history of Strepsiptera were broadly adaptations relating to the evolution to endoparasitism. These results are consistent with the hypothesis that parasitism may be an important cause of molecular evolutionary rate variation [31]–[33]. A plausible scenario could have involved deleterious mutations in free-living species becoming neutral/nearly-neutral in progressively host-dependent endoparasites. An adaptive interpretation could be that parasitism indirectly led to the selection of increased variation (through recombination or mutation), due to increased red-queen pressures between the host and parasite. But such a hypothesis does not explain why a subsequent decrease in substitution rate is observed in descendent nodes within Strepsiptera. Under the first (non adaptive) model, once most sites had been exposed to novel evolutionary forces, the substitution rate returned to a background level. Our findings are more in-keeping with relaxation of selective constraint as a dominating force in the early evolution of Strepsiptera.
However, the precise relationship between molecular rate and parasitism cannot be conclusively resolved in the current framework. Confounding correlates of endoparasitism may prove causally more relevant [2]. For example, in studies of mammalian molecular evolution, the increase in availability of DNA sequence data has questioned initial hypotheses positing a simple correlation with body size. Later studies used more reliable rate estimates and better methods to demonstrate that rate in the nuclear genome covaried with generation time and fecundity (and body size) but that variation in the rate of the mitochondrial genome was explained by longevity [43] (although in our study, rates between nuclear and mitochondrial genomes are similar). An alternative explanation in Strepsiptera could be that endoparasitism enabled females to increase individual fecundity by being able to concentrate more resources on one aspect of life-history: reproduction. Higher mutation rate could have subsequently stemmed from the associated increase in germline replications per generation. Endoparasitism may have also been correlated with increased generation time and shorter lifespan, where pressures to reproduce prior to host-death or clearance are considered to be critical components of parasite evolution [44]. These factors could be causally important in explaining the evolution of molecular rate in Strepsiptera, but determining which requires a more detailed understanding of life-history, ecology and the fine-scale interaction between Strepsiptera and host. Uncovering the mechanism of immune evasion could represent a particularly important target for future research.
The Strepsiptera long-branch
This study indicates that elevated molecular evolutionary rate was an important contributing cause of the strepsipteran long-branch. However, missing data in the form of undiscovered extinct (or extant) transitional lineages and imprecision over the nearest extant sister-lineage are also relevant to improving understanding of the causes of molecular rate variation in Strepsiptera. A number of recent studies consolidate the view that Strepsiptera are closely related to Coleoptera [27]–[30] but a precise hypothesis has still not been reached. Increased knowledge of strepsipteran life-history and ecology, in combination with a more detailed understanding of strepsipteran sister-relationships, will lead to better estimations of divergence, allowing for more informative date priors to be incorporated into a relaxed phylogenetic approach [45], [46]. Together, these will help to develop a more accurate picture of the forces responsible for variation in the rate of genome evolution in Strepsiptera. Revisiting hypotheses, like a possible association with rhipiphorine beetles [47] may help to identify potential candidate taxa that interrupt the branch. Alongside approaches that implement more sensitive phylogenetic methodology and larger data sets [48], new data may offer greater understanding of strepsipteran origins. However, this study suggests that the strepsipteran long-branch may never be easy to “break up”.
Conclusions
In this report, we present the first molecular phylogeny of Strepsiptera. Estimates of morphological branch length, alongside reconstruction of the strepsipteran phenotype reveal a correlation between morphological traits linked to endoparasitism and rate of molecular evolution. The main phase of diversification (Stylopidia, Stylopiformia) is associated with a return to a low and even rate of molecular evolution, and a period of relative morphological stability. This pattern supports the hypothesis that the transition to parasitism from a free-living insect ancestor can affect molecular rate. Greater precision over the nearest extant strepsipteran sister group will lead to better estimations of both divergence and molecular rate. Improved understanding of strepsipteran biology will in future permit the causes of molecular rate variation in Strepsiptera to be examined in greater detail. Together, these results establish an important foundation for further research into the evolution and ecology of a highly unusual host-parasite system.
Materials and Methods
Taxon and DNA sampling
Individuals were included from 41 strepsipteran taxa, across 16 genera (50% coverage). Bohartillidae and Bahiaxenidae, which are rare and represented by few specimens, were not included [49]. Three hemi- and six holometabolous outgroup species were selected from nucleotide data in Genbank. Due to the nature of mitochondrial gene evolution in Hymenoptera [50], [51] and the possibility of long-branch attraction between Strepsiptera and Hymenoptera (Hayward et al. in preparation; Figure 4 in [27]), the latter were not included. Specimens were preserved at 4°C in 95% ethanol, and protocols employed for sequence generation follow [8]. The mitochondrial genes cytochrome c oxidase I (cox1), NADH dehydrogenase I (nad1), and small subunit ribosomal RNA (16S rRNA), and nuclear gene small subunit ribosomal RNA (18S rRNA) were chosen to represent independent and variable evolutionary rates (Genbank accession JN082786–JN082922). Chromatograms were inspected manually using FinchTV (www.geospiza.com), and cox1 and nad1 fragments were aligned by eye in BioEdit [52], using translated nucleotides to guide the management of indels. 16S and 18S rRNA fragments were aligned manually using the comparative structural method [6], [53], [54] and mfold [55], but these do not correspond strictly to the category definitions sensu Gillespie [54].
A final alignment consisting of 3930 nucleotides was used in subsequent analyses, consisting of 967 bp 803 bp, and 2160 bp for the 18S rRNA, 16S rRNA, and cox1+nad1 partitions respectively, each with 339, 433, and 433 parsimony-informative positions. This approach was compared against an automated alignment strategy using the default settings in MUSCLE [56] and Gblocks [57], but retaining columns with a gap at greater than 50% of taxa. The resultant alignment contained 21% fewer characters (3048 nucleotides) of 642 bp, 774 bp and 1632 bp in the 18S rRNA, 16S rRNA and cox1+nad1 partitions, each with 229, 546, and 341 parsimony informative positions respectively. All analyses in this study are based on the structurally-informed “manual” alignment as trees based on the automated approach produced trees with limited support and equivocal topologies (data not shown). For the estimation of molecular rates, divergence estimates and date-informed branch lengths, the mitochondrial cox1 and nad1 genes were combined into a single data partition and analysed separately with/without the 3rd codon position (datasets MIT1+2 and MIT123 respectively). Specimen information (including accession numbers, primer information and 18S/16S rRNA template alignments) appears in Table S4.
Evolutionary model selection and phylogenetic analysis
For Bayesian analyses (BI), the most appropriate models of evolution were selected by comparing harmonic means across separate gene partitions in MrBayes v3.1.2 [58], [59], and then calculating Bayes Factor (BF) values. For maximum likelihood (ML), the Akaike Information Criterion (AIC) approach in MrModelTest v2 [60] and ProtTest v2.4 [61]–[63] was employed to select the most suitable models for RAxML v7.0.3 [64]. For all nucleotide partitions, the GTR+Γ+I model was preferred by BF and AIC with the following harmonic means: −7209.42 (18S rRNA); −9235.59 (16S rRNA); −14796.14 (MIT1+2); −30436.50 (MIT123) and log-likelihoods: −7160.8037; −9189.0898; −14758.0146; −31345.5293. For partition MIT123, the number of transitions and transversions estimated under the F84 model were plotted against genetic distance for each codon position using DAMBE v5.0.8 [65] (Figure S3, Table S5). A test of substitution saturation [66] and quartet likelihood mapping (TREE-PUZZLE v5.2) [67], [68] indicated high percentages of noise versus signal (20.6% and 32.7% in cox1 and nad1 respectively) in synonymous 3rd codon position, and little correspondence between 3rd codon position transition frequencies and genetic distance. Consequently, the mixed amino acid model facilitated by MrBayes was selected for use in all concatenated BI analyses. The amino acid substitution model favoured by the posterior density was MtRev [69] +Γ+I (+F). ProtTest found highest support for MtArt [70] and LG [71], but these are unavailable to MrBayes v3.1.2 and RAxML v7.0.3.
After models were selected, concatenated BI analyses consisted of two independent (MC)3 algorithms running for 2 million generations, each with four chains (3 hot, 1 cold), sampling one tree in 200, burn-in cutoffs were inspected manually for each parameter file in Tracer v1.4 [72]: the first 40000 steps were discarded. Inspection of the standard deviation of split frequencies confirmed that runs had converged (0.0059). All parameters except topology were unlinked between partitions. Data were summarized over a majority rule consensus tree (50% cutoff). 1000 ML nonparametric bootstrap pseudoreplicates were estimated in RAxML v7.0.3 [73]. 500 ML bootstrap pseudoreplicates without Lychnocolax were also estimated. Trees were imported into FigTree v1.2.3 for editing [74].
Divergence time estimation
MIT1+2 was employed in a Bayesian relaxed clock framework in BEAST v1.4.8 [75] using the GTR+Γ+I model. Lychnocolax taxa were removed prior to analyses and Holometabola was constrained as monophyletic. Likelihood ratio tests using least and most complex evolutionary models in PAML v4 [76], with/without the 3rd codon position were overdispersed with respect to a molecular clock (2ΔlnL = 1398.37, 1539.43, 836.45, 948.41; df = 7, P<0.001). Significant rate-heterogeneity was accommodated by employing the relaxed-clock MCMC with an uncorrelated lognormal model (UCLN) [77], calibrated using three strepsipteran fossils. The implementation of fossil priors is described in Text S1.
MCMC analyses ran for 10 million iterations, sampling every 1000th step. The effect of A+T-rich 3rd codon positions was investigated using the MIT123 dataset, in which the two partitions (1+2)(3) were unlinked. Analyses were repeated using the 18S rRNA dataset. The effect of model choice was assessed by comparing GTR+Γ+I with the SRD framework [78]. Molecular rate estimates were calculated as % pairwise sequence divergences per million years: equal to twice the per lineage rate. Dates were specified as millions of years before present, the Yule process was employed as the tree prior. Parameter files were inspected manually to ensure chain stability across parameters, and to select an appropriate burn-in. Tree files were summarized on a maximum clade credibility (MCC) tree.
Molecular rate, diversification rate, and tree shape
Relative rates of molecular evolution were examined via cross comparison of families and outgroups in RRTree v1.1.11 [79]. Whole-tree and single node methods were employed to test for departures in diversification rate [39]. The temporal rate cladogenesis test (RCT) statistic was calculated [80], [81]: nodes showing a “trickle-down” effect [39] were excluded. Whole tree simulations of rate-constant/rate-variable variants of the birth-death model in the Laser R-package [82] were conducted. The simulation ran for 1000 trees, comparing the best constant speciation model versus best variable speciation model (ΔAICrc) using the MIT1+2 tree. Outgroup taxa were pruned, and the birth rate (b), death rate (d) and unsampled taxa (m) were varied. m represents unsampled Strepsiptera species diversity. SymmeTree v1.1 [83] was used as an independent topological method. To investigate the impact of missing taxa, 560 tips were assigned to known groupings as soft polytomies. Whole-tree and single-node statistics were calculated using 100000 Bayesian simulations.
Evolutionary trait reconstruction
For reconstructions of morphology, data were imported into BayesTraits v1.0 [84]. Ancestral states were enforced using the ‘fossil’ prior. Harmonic means were compared for fossilized states, and accepted or rejected using BF values. 2 million MCMC iterations were conducted using the final consensus branching pattern and repeated if harmonic means did not stabilize. A reverse-jump hyperprior with exponential distribution 0–30 was set. ‘ratedev’ was optimized so that proposals were accepted 20–40% of the time. For host-use, major infraordinal divisions were treated as states. Aculeata (ants) were placed as the primary (ancestral) host for Myrmecolacidae: males of Myrmecolacidae parasitize only ants [85] and evidence for a female myrmecolacid in a fossilized ant host [86] indicated this was appropriate. A maximum likelihood approach, using the symmetrical method (Mesquite v2.71 [87]) was implemented to offer an independent measure of support.
Comparison of molecular and phenotypic rates of evolution
State changes corresponding to non-homoplasious steps from a morphological phylogeny of Strepsiptera [34] were mapped to shared nodes of the MIT1+2 MCC tree (excluding branches leading to missing taxa, conflicting nodes, and clades represented by one taxon – representing 6, 6 and 9 steps respectively). A smaller list of discrete adult and secondary larval characters was used in a Bayesian reconstruction approach to ensure this pattern was repeatable across methods.
As a formal comparison of the relationship between molecular and phenotypic evolution, morphological branch lengths were estimated in Phylip [88], using the genus-level matrix of adult and secondary larval characters from [34], updated to include the current set of taxa. The topology was constrained to follow the MIT1+2 dated MCC tree. For individual nodes, we tested the correlation between % pairwise sequence divergence and morphological branch length using standard statistics in R v2.9.2. As an independent whole-tree method, the K-score was calculated using Ktreedist v1.0 [89] and compared against a null K-distribution (500 simulated trees; following [18]). In this approach, molecular branch length (instead of pairwise sequence divergence) was assessed against morphological branch length.
Supporting Information
rRNA variabe and core domain structural attributes mapped onto the Strepsiptera phylogeny. (A) 18S variable (bar) and core (filled circle) A+T% content. (B) 16S variable (bar) and core (filled circle) A+T% content. (C) Variable domain size (nucleotide length) for the 18S (red) and 16S (black) genes. Outgroups grey and highlighted. Clade abbreviations and colour scheme follow Figure 1. Note the shifts in variable domain bp length, in both the 18S (length increase) and 16S (length decrease) genes at the node leading to Strepsiptera in (C).
(TIF)
Divergence time and molecular rate patterns using the nuclear 18S rRNA dataset. Red: % molecular rate mapped for each node at corresponding distances from root. Blue: Ln number of cumulative lineages at corresponding distances from root. This corroborates the analysis using the mitochondrial partition (Figure 2), confirming that the observed pattern is consistent across genomic compartments.
(TIF)
Exploration of data quality across the mitochondrial genes. Transitions and transversions estimated under the F84 model were plotted against genetic distance for each codon position: Green = 1sts, Blue = 2nds, Orange = 3rds. Signal versus noise was graphically visualized using quartet likelihood mapping.
(TIF)
Summary of Strepsiptera divergence times. Summary of divergence time estimates for the major nodes in the Strepsiptera phylogeny using the combined mitochondrial coding gene (cox1+nad1) partition. *Pairwise sequence divergences per million years. Clade abbreviations follow figure 1. †Node ages defined by exponential priors.
(DOC)
RRTest comparative analysis across strepsipteran clades. Bold = P-value with significant rate comparison (bonferroni corrected). *Marginally non-significant after bonferroni adjustment in the mitochondrial (A) and 18S rRNA partition (B). Clade abbreviations follow figure 1.
(DOC)
List of characters and corresponding states recovered in the reconstruction of strepsipteran morphological traits. The position in the phylogeny of significant character reconstructions appears in brackets next to the corresponding state, followed by the BF range supporting that reconstruction. Some characters may be considered dependent, if single genotypic events can be demonstrated to produce pleiotropic effects. Possible examples include male/female larval spiracles, and male/female larval legs. * equivocal BFs (0.2–3.8). This might disguise a potentially apomorphic loss of tarsomeres in the Elenchidae+Halictophagidae ancestor.
(DOC)
Specimen, primer information and rRNA template alignments. Genbank accession and specimen source information; list of primers used in this study (*Primers designed for short-fragment PCR) and 18 rRNA and 16S rRNA template secondary structural alignments.
(DOC)
Test of saturation by mitochondrial gene and codon position. *Statistics indicating little saturation. †Statistics with substantial saturation (bold). ‡Statistics indicating useless/very poor sequence for phylogenetics (bold). Ts = symmetrical T-statistic. Tns = non-symmetrical T-statistic.
(DOC)
(DOC)
Acknowledgments
We thank Peter W. H. Holland for use of laboratory facilities; St Hugh's College, Oxford; the National Museum of Natural History; the Smithsonian Institution (Washington DC) and the Smithsonian Tropical Research Institute (Panama) for support to JK. We thank three anonymous reviewers and Jordi Paps for helpful suggestions. We are indebted to many collectors for providing invaluable material (listed in Text S1).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by The Leverhulme Trust (F/08 502/G). DPM was supported by an Elizabeth Hannah Jenkinson Fund for laboratory funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.O'Conner TD, Mundy NI. Genotype-phenotype associations: substitution models to detect evolutionary associations between phenotypic variables and genotypic evolutionary rate. Bioinformatics. 2009;25:i94–i100. doi: 10.1093/bioinformatics/btp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanfear R, Welch JJ, Bromham L. Watching the clock: studying variation in rates of molecular evolution between species. Trends Ecol Evol. 2010;25:495–503. doi: 10.1016/j.tree.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Kathirithamby J. Review of the order Strepsiptera. Syst Entomol. 1989;14:41–62. [Google Scholar]
- 4.Kathirithamby J. Host-parasitoid associations in Strepsiptera. Annu Rev Entomol. 2009;54:227–249. doi: 10.1146/annurev.ento.54.110807.090525. [DOI] [PubMed] [Google Scholar]
- 5.Kathirithamby J, Ross LD, Johnston JS. Masquerading as self? endoparasitic Strepsiptera (Insecta) enclose themselves in host-derived epithelial bag. P Natl Acad Sci USA. 2003;100:7655–7659. doi: 10.1073/pnas.1131999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillespie JJ, McKenna CH, Yoder MJ, Gutell RR, Johnston JS, et al. Assessing the odd secondary structural properties of nuclear small subunit ribosomal RNA sequences (18S) of the twisted-wing parasites (Insecta: Strepsiptera). Insect Mol Biol. 2005;14:625–643. doi: 10.1111/j.1365-2583.2005.00591.x. [DOI] [PubMed] [Google Scholar]
- 7.Pohl H, Beutel RG. The evolution of Strepsiptera. Zoology. 2007;111:318–338. doi: 10.1016/j.zool.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 8.McMahon DP, Hayward A, Kathirithamby J. The mitochondrial genome of the ‘twisted-wing parasite’ Mengenilla australiensis: a comparative study. BMC Genomics. 2009;10:603. doi: 10.1186/1471-2164-10-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter MS, Woolley JB. Evolution and behavioural ecology of heteronomous aphelinid parasitoids. Annu Rev Entomol. 2001;46:251–90. doi: 10.1146/annurev.ento.46.1.251. [DOI] [PubMed] [Google Scholar]
- 10.Hayward A, McMahon DP, Kathirithamby J. Cryptic diversity and host specificity in a parasitoid where the sexes utilize hosts from separate orders. Mol Ecol. 2011;20:1508–1528. doi: 10.1111/j.1365-294X.2011.05010.x. [DOI] [PubMed] [Google Scholar]
- 11.Eggleton P, Belshaw R. Insect parasitoids: an evolutionary overview. Philos Trans R Soc London Ser B. 1992;337:1–20. [Google Scholar]
- 12.Kinzelbach RK. Strepsiptera. Die Tierwelt Deutschlands. 1978;65:166. [Google Scholar]
- 13.Poulin R, Morand S. Parasite biodiversity. Washington DC: Smithsonian Institution Press; 2004. [Google Scholar]
- 14.Poulin R, Keeney DB. Host specificity under molecular and experimental scrutiny. Trends Parasitol. 2008;24:24–28. doi: 10.1016/j.pt.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT. Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat Genet. 2004;36:1326–1329. doi: 10.1038/ng1471. [DOI] [PubMed] [Google Scholar]
- 16.Nadeau NJ, Burke T, Mundy NI. Evolution of an avian pigmentation gene correlates with a measure of sexual selection. Proc R Soc B Biol Sci. 2007;255:37–45. doi: 10.1098/rspb.2007.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramm SA, Oliver PL, Ponting CP, Stockley P, Emes RD. Sexual selection and the adaptive evolution of mammalian ejaculate proteins. Mol Biol Evol. 2008;25:207–219. doi: 10.1093/molbev/msm242. [DOI] [PubMed] [Google Scholar]
- 18.Ahrens D, Ribera I. Inferring speciation modes in a clade of Iberian chafers from rates of morphological evolution in different character systems. BMC Evol Biol. 2009;9:234. doi: 10.1186/1471-2148-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H, Lee W, Lee S. Morphometric relationship, phylogenetic correlation, and character evolution in the species-rich genus Aphis (Hemiptera: Aphididae). PLoS ONE. 2010;5:e11608. doi: 10.1371/journal.pone.0011608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardman M, Hardman LM. The relative importance of body size and paleoclimatic change as explanatory variables influencing lineage diversification rate: an evolutionary analysis of bullhead catfishes (Siluriformes: Ictaluridae). Syst Biol. 2008;57:116–130. doi: 10.1080/10635150801902193. [DOI] [PubMed] [Google Scholar]
- 21.Omland KE. Correlated rates of molecular and morphological evolution. Evolution. 1997;51:1381–1393. doi: 10.1111/j.1558-5646.1997.tb01461.x. [DOI] [PubMed] [Google Scholar]
- 22.Bromham L, Woolfit M, Lee MSY, Rambaut A. Testing the relationship between morphological and molecular rates of change along phylogenies. Evolution. 2002;56:1921–1930. doi: 10.1111/j.0014-3820.2002.tb00118.x. [DOI] [PubMed] [Google Scholar]
- 23.Ekman S, Andersen HL, Wedin M. The limitations of ancestral state reconstruction and the evolution of the Ascus in the Lecanorales (lichenized Ascomycota). Syst Biol. 2007;57:141–156. doi: 10.1080/10635150801910451. [DOI] [PubMed] [Google Scholar]
- 24.Whiting MF, Carpenter JC, Wheeler QD, Wheeler WC. The Strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Syst Biol. 1997;46:1–68. doi: 10.1093/sysbio/46.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Huelsenbeck JP. Systematic bias in phylogenetic analysis: is the Strepsiptera problem solved? Syst Biol. 1998;47:519–537. [PubMed] [Google Scholar]
- 26.Huelsenbeck JP. A Bayesian perspective of the Strepsiptera problem. Tidjschr Ent. 2001;144:165–178. [Google Scholar]
- 27.Wiegmann BM, Trautwein MD, Kim JW, Cassel BK, Bertone MA, et al. Single-copy nuclear genes resolve the phylogeny of the holometabolous insects. BMC Biology. 2009;7:34. doi: 10.1186/1741-7007-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longhorn SJ, Pohl H, Vogler AP. Ribosomal protein genes of holometabolous insects reject the Halteria, instead revealing a close affinity of Strepsiptera with Coloeptera. Mol Phylogenet Evol. 2010;55:846–859. doi: 10.1016/j.ympev.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 29.Mckenna DD, Farrell BD. 9-genes reinforce the phylogeny of Holometabola and yield alternative views on the phylogenetic placement of Strepsiptera. PLoS ONE. 2010;5:e11887. doi: 10.1371/journal.pone.0011887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishiwata K, Saski G, Owaga J, Miyata T, Su ZH. Phylogenetic relationships among insect orders based on three nuclear protein-coding gene sequences. Mol Phylogenet Evol. 2010 doi: 10.1016/j.ympev.2010.11.001. (doi:10.1016/j.ympev.2010.11.001) [DOI] [PubMed] [Google Scholar]
- 31.Bromham L. Why do species vary in their rate of molecular evolution? Biol Lett. 2009;5:401–404. doi: 10.1098/rsbl.2009.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dowton M, Austin AD. Increased genetic diversity in mitochondrial genes is correlated with the evolution of parasitism in the Hymenoptera. J Mol Evol. 1995;41:958–965. doi: 10.1007/BF00173176. [DOI] [PubMed] [Google Scholar]
- 33.Duff RJ, Nickrent DL. Characterization of mitochondrial small-subunit ribosomal RNAs from holoparasitic plants. J Mol Evol. 1997;45:631–639. doi: 10.1007/pl00006267. [DOI] [PubMed] [Google Scholar]
- 34.Pohl H, Beutel RG. The phylogeny of Strepsiptera (Hexapoda). Cladistics. 2005;21:328–374. doi: 10.1111/j.1096-0031.2005.00074.x. [DOI] [PubMed] [Google Scholar]
- 35.Brower AVZ. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. P Natl Acad Sci USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrell BD. Evolutionary assembly of the milkweed fauna: cytochrome oxidase I and the age of Tetraopes beetles. Mol Phylogenet Evol. 2001;18:46–478. doi: 10.1006/mpev.2000.0888. [DOI] [PubMed] [Google Scholar]
- 37.Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 38.Ricklefs RE, Losos JB, Townsend TM. Evolutionary diversification of clades of squamate reptiles. J Evol Biol. 2007;20:1751–1762. doi: 10.1111/j.1420-9101.2007.01388.x. [DOI] [PubMed] [Google Scholar]
- 39.Silvestri F. Studi sugli ‘Strepsiptera’ Insecta. III. Descrizione e biologia di 6 specie italiane di Mengenilla. Boll Lab Zool Gen Agric Portici. 1943;32:197–282. [Google Scholar]
- 40.Pohl H, Beutel RG. Fine structure of adhesive devices of Strepsiptera (Insecta). Arthropod Struct Dev. 2004;33:31–43. doi: 10.1016/j.asd.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Moore BR, Donoghue MJ. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovation. Am Nat. 2007;170:S28–S55. doi: 10.1086/519460. [DOI] [PubMed] [Google Scholar]
- 42.Venditti C, Meade A, Pagel M. Phylogenies reveal new interpretation of speciation and the red queen. Nature. 2009;463:349–352. doi: 10.1038/nature08630. [DOI] [PubMed] [Google Scholar]
- 43.Welch JJ, Bininda-Emonds ORP, Bromham L. Correlates of substitution rate variation in mammalian protein-coding sequences. BMC Evol Biol. 2008;8:53. doi: 10.1186/1471-2148-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid-Hempel P. Parasite immune evasion: a momentous molecular war. Trends Ecol Evol. 2008;23:318–26. doi: 10.1016/j.tree.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 45.Welch JJ, Bromham L. Molecular dating when rates vary. Trends Ecol Evol. 2005;20:320–327. doi: 10.1016/j.tree.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 46.Ho SYW. An examination of phylogenetic models of substitution rate variation among lineages. Biol Lett. 2009;5:421–424. doi: 10.1098/rsbl.2008.0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crowson RA. The phylogeny of Coleoptera. Ann Rev Entomol. 1960;5:111–134. [Google Scholar]
- 48.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino acid replacement process. Mol Biol Evol. 2004;21:1095–109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 49.Bravo F, Pohl H, Silvo-Neto A, Beutel RG. Bahiaxenidae, a “living fossil” and a new family of Strepsiptera (Hexapoda) discovered in Brazil. Cladistics. 2009;25:1–10. doi: 10.1111/j.1096-0031.2009.00264.x. [DOI] [PubMed] [Google Scholar]
- 50.Castro LR, Dowton M. The position of the Hymenoptera within the Holometabola as inferred from the mitochondrial genome of Perga condei (Hymenoptera: Symphyta: Pergidae). Mol Phylogenet Evol. 2005;34:469–479. doi: 10.1016/j.ympev.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Castro LR, Dowton M. Mitochondrial genomes in the Hymenoptera and their utility as phylogenetic markers. Syst Entomol. 2006;32:60–69. [Google Scholar]
- 52.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 53.Kjer KM, Baldridge GD, Fallon AM. Mosquito large subunit ribosomal RNA: simultaneous alignment of primary and secondary structure. Biochim Biophys Acta. 1994;1217:147–155. doi: 10.1016/0167-4781(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 54.Gillespie JJ. Characterizing regions of ambiguous alignment caused by the expansion and contraction of hairpin-stem loops in ribosomal RNA molecules. Mol Phylogenet Evol. 2004;33:936–943. doi: 10.1016/j.ympev.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Talavera G, Castresana J. Improvement of phylogenies after removing divergent ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 58.Huelsenbeck J, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 59.Ronquist F, Huelsenbeck JP. MrBayes 3, Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 60.Nylander JAA. MrModeltest v2. 2004. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- 61.Drummond A, Strimmer K. PAL: An object-oriented programming library for molecular evolution and phylogenetics. Bioinformatics. 2001;17:662–663. doi: 10.1093/bioinformatics/17.7.662. [DOI] [PubMed] [Google Scholar]
- 62.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 63.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 64.Stomatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 65.Xia X, Xie Z. DAMBE: data analysis in molecular biology and evolution. J Heredity. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 66.Xia X, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Mol Phylogenet Evol. 2003;26:1–7. doi: 10.1016/s1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- 67.Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 69.Adachi J, Hasegawa M. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol. 1996;42:459–468. doi: 10.1007/BF02498640. [DOI] [PubMed] [Google Scholar]
- 70.Abascal F, Posada D, Zardoya R. MtArt: A new model of amino acid replacement for Arthropoda. Mol Biol Evol. 2007;24:1–5. doi: 10.1093/molbev/msl136. [DOI] [PubMed] [Google Scholar]
- 71.Le SQ, Gascuel O. LG: an improved, general amino-acid replacement matrix. Mol Biol Evol. 2008;25:1307–20. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 72.Rambaut A, Drummond AJ. 2007. Tracer v1.4, Available from http://beast.bio.ed.ac.uk/Tracer.
- 73.Stomatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 74.Rambaut A, Drummond AJ. 2007. (2007) FigTree v1.0, Available from http://tree.bio.ed.ac.uk/FigTree.
- 75.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Z. PAML 4, a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 77.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- 79.Robinson-Rechavi M, Huchon D. RRTree: Relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics. 2000;16:296–297. doi: 10.1093/bioinformatics/16.3.296. [DOI] [PubMed] [Google Scholar]
- 80.Nee S, Barraclough TG, Harvey PH. Temporal changes in biodiversity: detecting patterns and identifying causes. In: Gaston KJ, editor. Biodiversity: a biology of numbers and differences. Oxford: Blackwell Science; 1996. pp. 230–252. [Google Scholar]
- 81.Rambaut A, Harvey PH, Nee S. End-Epi: an application for inferring phylogenies and population dynamic processes from molecular sequences. Comput Appl Biosci. 1997;13:303–306. doi: 10.1093/bioinformatics/13.3.303. [DOI] [PubMed] [Google Scholar]
- 82.Rabosky DL. Likelihood methods for inferring temporal shifts in diversification rates. Evolution. 2006;60:1152–1164. [PubMed] [Google Scholar]
- 83.Chan KM, Moore BR. SYMMETREE: whole-tree analysis of differential diversification rates. Bioinformatics. 2005;21:1709–1710. doi: 10.1093/bioinformatics/bti175. [DOI] [PubMed] [Google Scholar]
- 84.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 85.Kathirithamby J, Hayward A, McMahon DP, Ferreira RS, Andreazze R, et al. Conspecifics of a heterotrophic heteronomous species of Strepsiptera (Insecta) are matched by molecular characterization. Syst Entomol. 2010;35:234–242. [Google Scholar]
- 86.Pohl H, Kinzelbach KJ. First record of a female stylopid (Strepsiptera: ?Myrecolacidae) parasite of prionomyrmecine ant (Hymenoptera: Formicidae) in Baltic amber. Insect Syst Evol. 2001;32:143–146. [Google Scholar]
- 87.Maddison WP, Maddison DR. Mesquite, a modular system for evolutionary analysis, Version 1.12. 2006. Available at: http://mesquiteproject.org.
- 88.Felsenstein J. PHYLIP-phylogeny inference package (version 3.2). Cladistics. 1989;5:164–166. [Google Scholar]
- 89.Soria-Carrasco V, Talavera G, Idea J, Castresana J. The K tree score: quantification of differences in the relative branch length and topology of phylogenetic trees. Bioinformatics. 2007;23:2954–2956. doi: 10.1093/bioinformatics/btm466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
rRNA variabe and core domain structural attributes mapped onto the Strepsiptera phylogeny. (A) 18S variable (bar) and core (filled circle) A+T% content. (B) 16S variable (bar) and core (filled circle) A+T% content. (C) Variable domain size (nucleotide length) for the 18S (red) and 16S (black) genes. Outgroups grey and highlighted. Clade abbreviations and colour scheme follow Figure 1. Note the shifts in variable domain bp length, in both the 18S (length increase) and 16S (length decrease) genes at the node leading to Strepsiptera in (C).
(TIF)
Divergence time and molecular rate patterns using the nuclear 18S rRNA dataset. Red: % molecular rate mapped for each node at corresponding distances from root. Blue: Ln number of cumulative lineages at corresponding distances from root. This corroborates the analysis using the mitochondrial partition (Figure 2), confirming that the observed pattern is consistent across genomic compartments.
(TIF)
Exploration of data quality across the mitochondrial genes. Transitions and transversions estimated under the F84 model were plotted against genetic distance for each codon position: Green = 1sts, Blue = 2nds, Orange = 3rds. Signal versus noise was graphically visualized using quartet likelihood mapping.
(TIF)
Summary of Strepsiptera divergence times. Summary of divergence time estimates for the major nodes in the Strepsiptera phylogeny using the combined mitochondrial coding gene (cox1+nad1) partition. *Pairwise sequence divergences per million years. Clade abbreviations follow figure 1. †Node ages defined by exponential priors.
(DOC)
RRTest comparative analysis across strepsipteran clades. Bold = P-value with significant rate comparison (bonferroni corrected). *Marginally non-significant after bonferroni adjustment in the mitochondrial (A) and 18S rRNA partition (B). Clade abbreviations follow figure 1.
(DOC)
List of characters and corresponding states recovered in the reconstruction of strepsipteran morphological traits. The position in the phylogeny of significant character reconstructions appears in brackets next to the corresponding state, followed by the BF range supporting that reconstruction. Some characters may be considered dependent, if single genotypic events can be demonstrated to produce pleiotropic effects. Possible examples include male/female larval spiracles, and male/female larval legs. * equivocal BFs (0.2–3.8). This might disguise a potentially apomorphic loss of tarsomeres in the Elenchidae+Halictophagidae ancestor.
(DOC)
Specimen, primer information and rRNA template alignments. Genbank accession and specimen source information; list of primers used in this study (*Primers designed for short-fragment PCR) and 18 rRNA and 16S rRNA template secondary structural alignments.
(DOC)
Test of saturation by mitochondrial gene and codon position. *Statistics indicating little saturation. †Statistics with substantial saturation (bold). ‡Statistics indicating useless/very poor sequence for phylogenetics (bold). Ts = symmetrical T-statistic. Tns = non-symmetrical T-statistic.
(DOC)
(DOC)