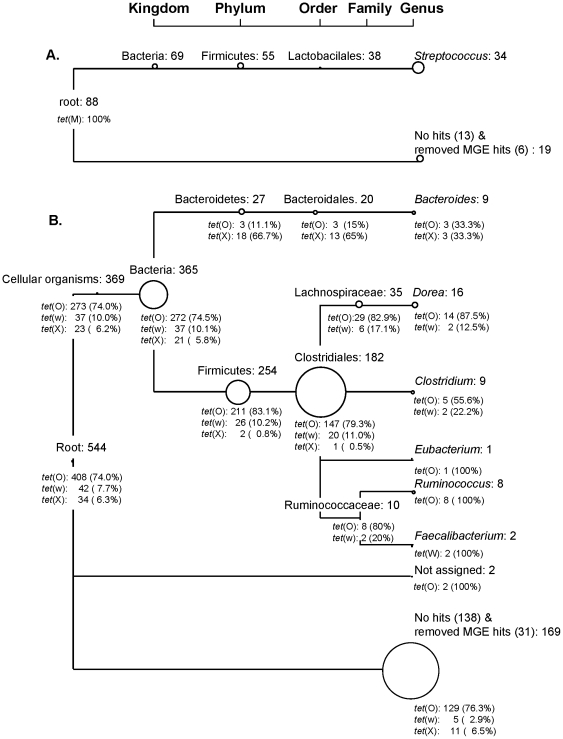
Figure 2. Microbial diversity of Tcr fosmid clones in infant (A) and mother (B) metagenomes.
Modified MEGAN tree (collapsed at Genus level) showing summarized number of reads assigned at different taxonomical levels. The size of each node is proportional to the number of reads assigned to the node. Beneath each node the number and percentage of Tcr genes detected in this study are noted. A. The “No hits & removed MGE hits” category contains 13 reads with no BLASTX hits (or hits that did not attain the min score/length of 0.15), 2 removed reads which were predicted to be located in MGE and were initially assigned below order level and 4 reads that mapped to ORFs in the sequenced transposon (Tn6079). B. The “No hits & removed MGE hits” category contains 138 reads with no BLASTX hits (or hits that did not attain the min score/length of 0.15) and 31 removed reads which were predicted to be located in MGE and were initially assigned below order level. The “Not assigned” category contains 2 reads that were assigned by BLASTX hits to uncultured bacteria.