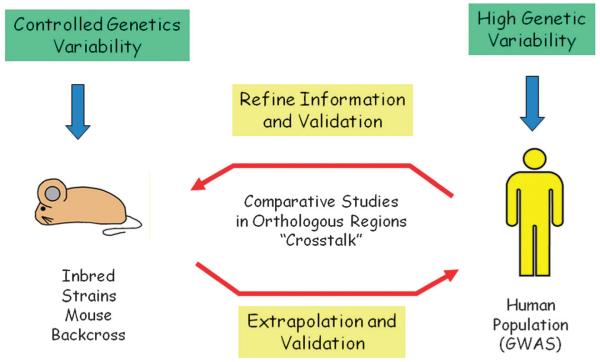
Fig. 4.
Mouse models are a good tool to identify QTLs in an environmentally controlled way that could be extrapolated to the human population. At the same time, they are very useful to verify and refine candidate loci found in humans by GWAS. Technical advances such as whole genome sequencing are readily making the recognition of orthologous chromosomal regions between species straight forward, simplifying the refinement of QTLs found in both kinds of studies.