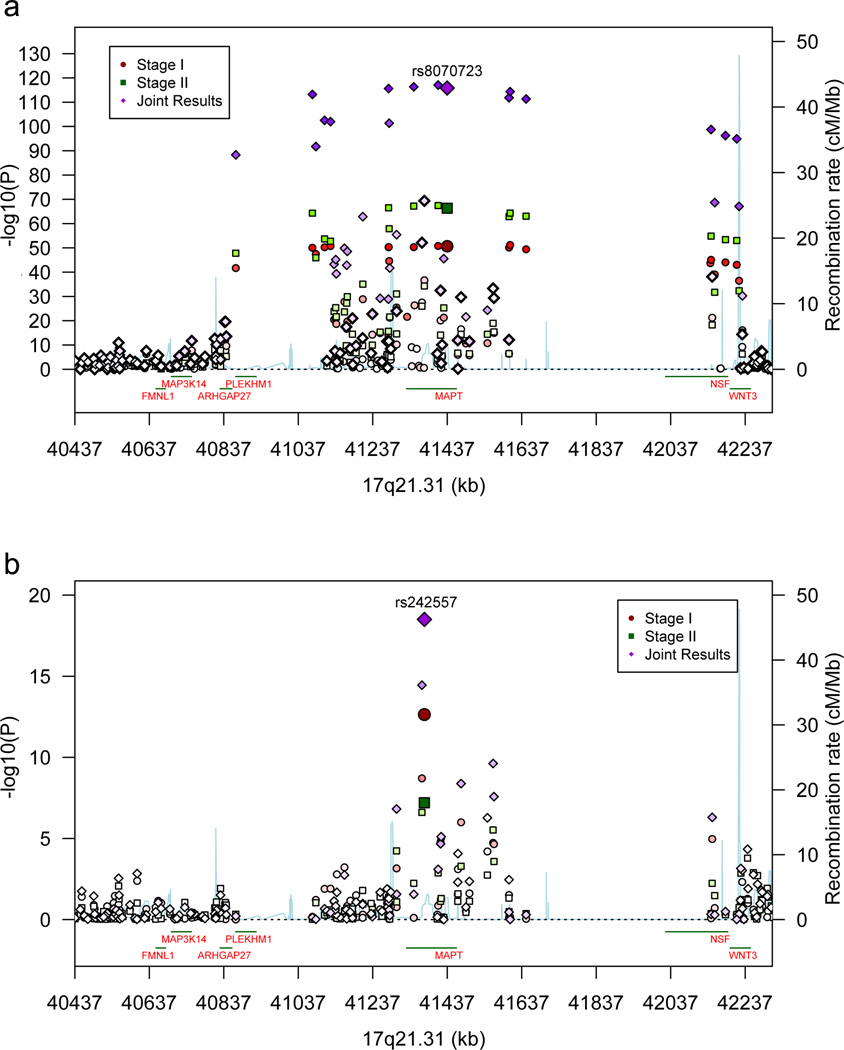
Figure 2.
(a) Association results for the 17q21.31 H1/H2 inversion polymorphism (40,974,015 – 41,926,692 Kb) and flanking segments. (b) Association results for 17q21.31 controlling for H1/H2. Results are shown for Stages 1 and 2 and the joint analyses. Recombination rate, calculated from the linkage disequilibrium (LD) structure of the region, is derived from Hapmap3 data. LD, encoded by intensity of the colors, is the pairwise LD of the most highly associated SNP at Stage 1 with each of the SNPs in the region.