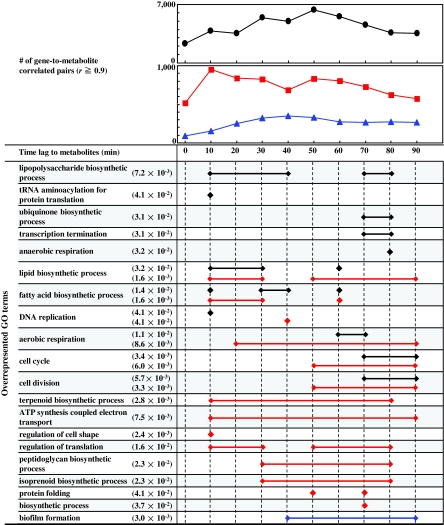
FIG. 2.
Gene-to-metabolite correlation analysis considering time lag between transcriptomics and metabolomics data. The plot shows the number of gene-to-metabolite (top: black), gene-to-unsaturated phosphatidylglycerols (PGs) (middle: red), and gene-to-cycropropanated PGs (middle: blue) correlated pairs (PCC ≥0.9) using 1,162 genes and 174 metabolites, respectively. Time lag of abscissa shows considered time lag between transcriptomics and metabolomics data. GO terms with FDR corrected p-value ≤ 0.05 (minimum value is indicated, if any) are indicated at the left side. Diamond and bar correspond to overrepresented GO terms by Fisher's exact test. lipopolysaccharide biosynthetic process is, for example, overrepresented in correlated pairs between transcriptomics and 10-, 20-, 30-, 40-, 70-, and 80-min time-lagged metabolomics data.