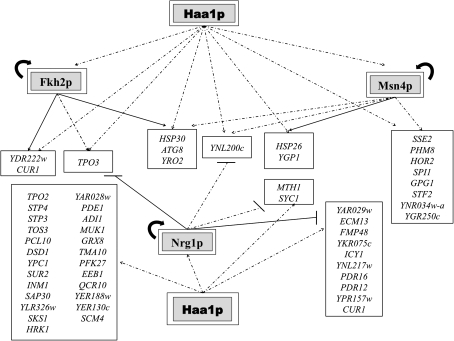
FIG. 8.
Transcriptional regulatory associations that may be governing the Haa1p-dependent response in yeast cells under acetic acid stress. The model is based on the microarray data obtained in this study and on the information available in the YEASTRACT database (December 2009). Genes whose transcription is activated in response to acetic acid in a Haa1p-dependent manner were selected for this analysis. The regulatory network is based on the documented regulatory associations between transcription factors (TF) whose expression is activated by Haa1p (the encoding genes are shown inside double boxes) and the other genes regulated by Haa1p, according with the microarray analysis carried out. Positive regulations between activators and target genes are represented by an arrow (→) and negative regulations between repressors and target genes are represented by lines (—). Documented associations are based on immunoprecipitation experiments (closed arrows or lines; → or —) or on transcription analysis approaches with no demonstration of the binding of the TF to the promoter region of target genes (dashed arrows or lines, -- -▸ or - · - · -).