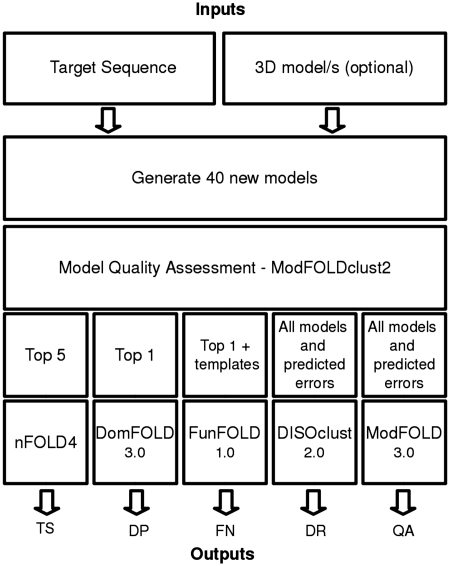
Figure 1.
Diagram of the software stack implemented for the IntFOLD server. This figure highlights the interdependency of the IntFOLD algorithms, with ModFOLDclust2 acting as the key algorithm in the IntFOLD pipeline. A query protein sequence is first submitted, with ∼40 new models generated. Secondly, the models are fed into the ModFOLDclust2 (7) model quality assessment algorithm, which ranks the models by model quality. The models are then used along with a combination of local error and template information to produce all the resulting output for 3D structure prediction (TS), domain prediction (DP), binding site residue prediction function prediction (FN), disorder prediction (DR) and model quality assessment (QA).