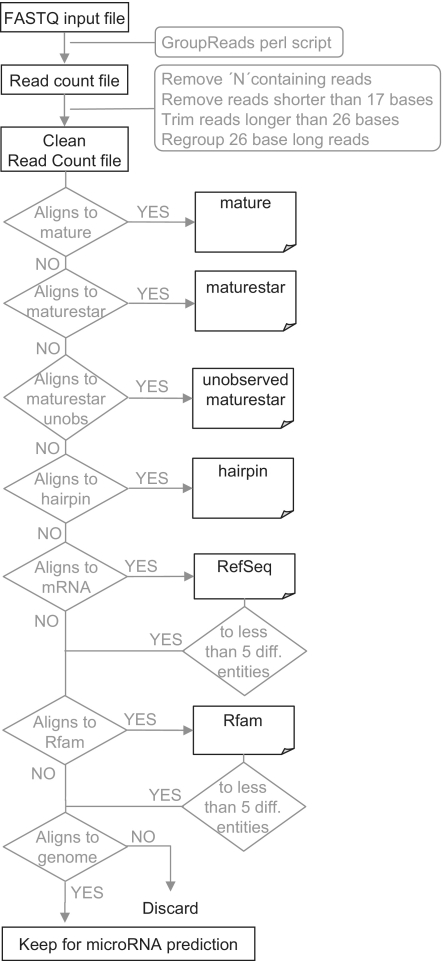
Figure 1.
General workflow of miRanalyzer. The fastq file is transformed into a read count file, which is filtered to keep only sequences from 17 to 26 bases. These reads are successively mapped to several databases in order to identify known microRNAs, discard messenger RNA contaminations and select sequences for the microRNA prediction step.