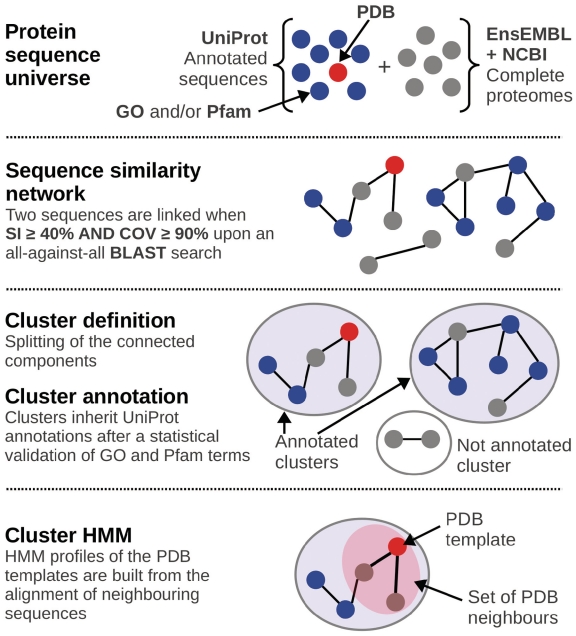
Figure 1.
BAR+ implementation. Our method collects sequences from the protein universe (UniProtKB) including also some 988 genomes. By this, all the features [PDB (± SCOP classification) (red circles), GO terms (including Molecular Function, Biological Process and Cellular Localization) and Pfam models (blue circles) are also included. An extensive BLAST alignment is performed of all the 13 495 736 sequences in a GRID environment. The sequence similarity network is built by connecting two sequences only if their SI is ≥40% with an overlapping COV ≥ 90%. About 913 762 clusters are obtained by splitting of the connected components. By this, any cluster may contain from 2 up to 87 893 sequences (one cluster containing ABC transporters from Prokaryotes, Eukaryotes and Archaea). Stand alone sequences are called Singletons (30.4% of the total protein universe). Sequences inherit the annotations within a cluster. When clusters are endowed with PDB template/s, a Cluster-HMM is generated by considering all the sequences that have an identity ≥ 40% and a COV ≥ 90% with the structure/s (pink subset). The Cluster-HMM can be used to align all the other sequences in the cluster to template/s.