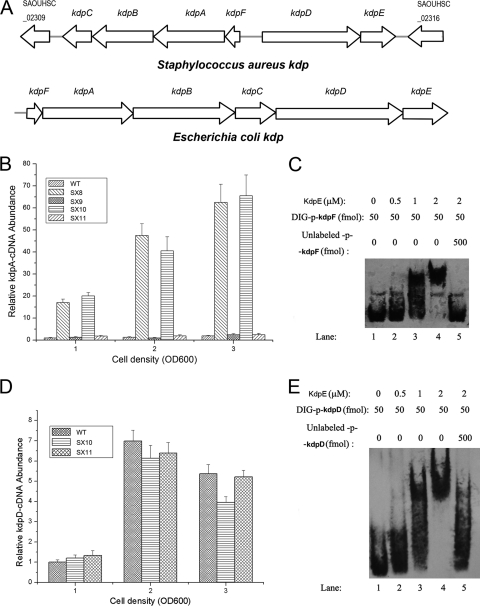
Fig. 1.
Regulatory effect of KdpE on the kdp operon and kdpDE transcription. (A) Organization of the kdp operons in S. aureus and E. coli. The arrows indicate the directions of translation as determined from the nucleotide sequence. (B) The regulatory effect of KdpDE on the transcription of kdpFABC in cells grown in LB medium. The transcript levels of kdpFABC were compared using real-time RT-PCR in wild type (WT; S. aureus NCTC8325), SX8 (kdpDE mutant), SX9 (kdpDE mutant with a plasmid encoding KdpDE), SX10 (kdpE mutant), and SX11 (kdpE mutant with a plasmid encoding KdpE). The strains were grown in LB medium to OD600s of 1, 2, and 3. (C) The ability of KdpE to bind to the kdpFABC promoter as determined by EMSAs. (D) The regulatory effect of KdpE on the transcription of kdpD. The transcript levels of kdpD were compared between WT, SX10 (kdpE mutant), and SX11 (kdpE mutant with a plasmid encoding KdpE). (E) The ability of KdpE to bind to the kdpDE promoter as determined by EMSAs. All the real-time PCR assays were repeated five times with similar results. Error bars indicate standard deviations.