Figure 3.
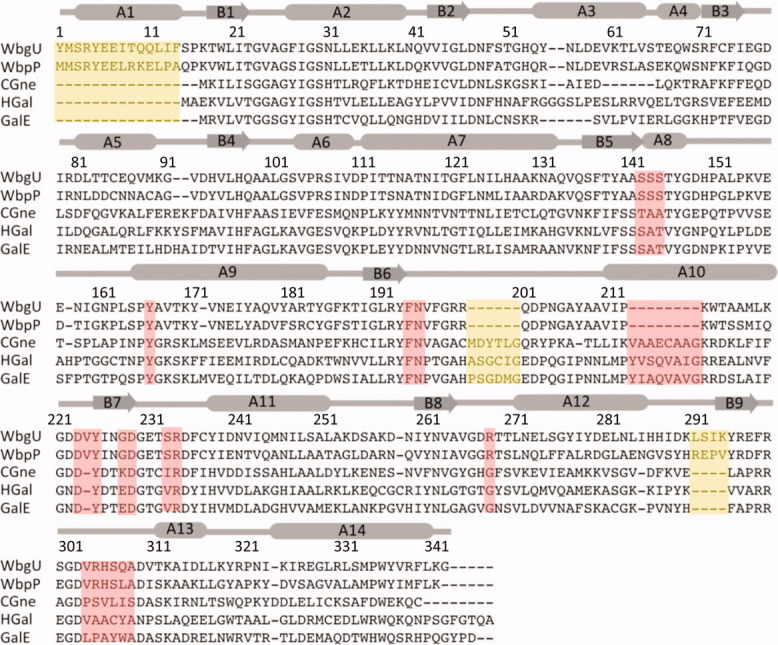
Multiple sequence alignment between 5 different UDP-hexose 4-epimerases. Regions deemed important for determination of substrate specificity are highlighted in pink. Regions highlighted in yellow are distinct structural variations that do not have a direct bearing on substrate binding or catalysis. WbgU is UDP-GalNAc 4-epimerase from P. shigelloides; WbpP is UDP-GlcNAc 4-epimerase from P. aeruginosa; CGne is UDP-Glc/GlcNAc 4-epimerase (Gne) from C. jejuni; HGal is UDP-Glc/GlcNAc 4-epimerase from Homo sapiens; GalE is UDP-Glc 4-epimerase from E.coli. The numbering and the secondary structure assignment corresponds the structure of WbgU. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
