Figure 5.
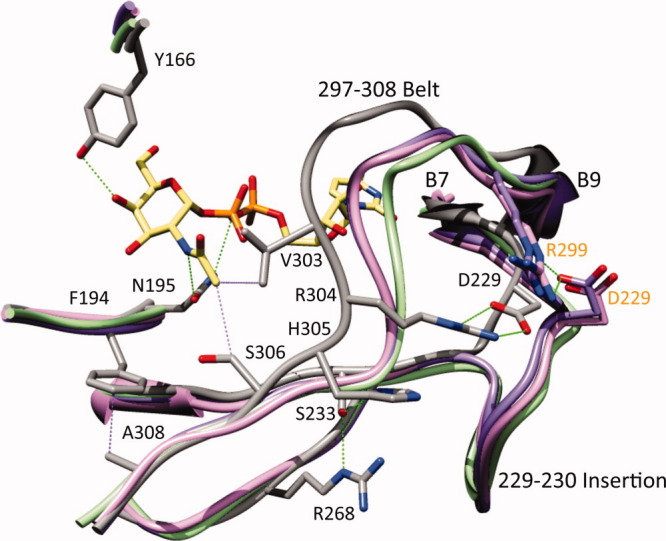
Substrate recognition by the group 1 and the group 2 UDP-hexose 4-epimerases from the perspective of the group 3. His305 in WbgU is substituted by Val in CGne and by Ala in GalE and HGal; Ser233 in WbgU is substituted by Ile in CGne and by Val in GalE and HGal; Arg268 in WbgU is substituted by Gly in CGne, HGal, and GalE (side chains are not shown for clarity). Similarly Arg304 is conserved in WbgU and WbpP whereas being substituted by Ser in CGne, Ala in HGal and Pro in GalE. The loss of the hydrogen bonding network at His305 position and the salt bridge at Arg304 position result in an altered conformation of substrate binding loop in the Group 1 and the Group 2 epimerases. In addition, the insertion of an 8 residue loop at the Asp229 causes formation of a salt bridge with Arg299. The absence of the polar interactions in the Group 1 and the Group 2 epimerases at the 304 and 305 positions in combination with the presence of a salt bridge at the 8 residue insertion between Asp229-Arg299, thus dictates the conformation of the substrate binding region in the group 1 and the group 2 epimerases. The common architecture of the substrate binding region in the group 1 and the group 2 epimerases has one important variation: Ser306 of WbgU (and WbpP) is substituted by a Cys in the Group 2 and by a Tyr in the group 1 hence restricting the access of bulky N-acetyl group to the active site of the group 1 epimerases, which in turn makes them specific towards the nonacetylated substrates. The labels and numbering in black color correspond to WbgU; the labels and numbering in orange color correspond to HGal. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
