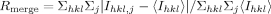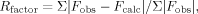Table I.
Data Collection and Refinement Statistics
| Data collection statistics | |
| Source | Beamline 9-2 SSRL |
| Space group | P32 |
| Unit cell parameters | |
| a, b, c (Å) | 78.1, 78.1, 231.9 |
| α, β, γ (deg) | 90, 90, 120 |
| Temperature (K) | 103 |
| Wavelength (Å) | 0.97 |
| Resolution (Å) | 44.03–2.50 (2.56–2.50)a |
| Rmergeb | 0.13 (0.9) |
| Completeness (%) | 94.7 (88.8) |
| 〈I/σ(I)〉 | 13.5 (1.4) |
| Number of unique reflections | 51939 (3563) |
| Average redundancy | 4.3 (4.1) |
| B factor from Wilson plot (Å2) | 44.9 |
| PDB entry | 3LU1 |
| Refinement statistics | |
| Resolution (Å) | 44.03-2.50 |
| Number of reflections | 51878 |
| Rfactorc | 20.7 |
| Rfreec | 25.8 |
| Number of protein atoms | 10684 |
| Number of water atoms | 182 |
| Number of ligand atoms | 332 |
| RMS deviation from ideal values for bond distances (Å)d | 0.01 |
| RMS deviation from ideal values for bond angles (deg)d | 1.15 |
| Average B-factors | |
| Main chain (Å2) | 36.6 |
| Side chain and waters (Å2) | 37.0 |
| All atoms (Å2) | 36.8 |
| Ramachandran plote | |
| Favored (%) | 95.8 |
| Disallowed (%) | 0.3 |