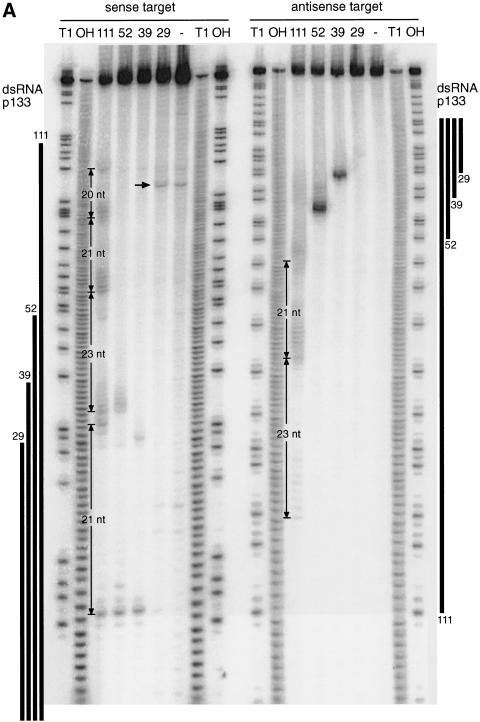
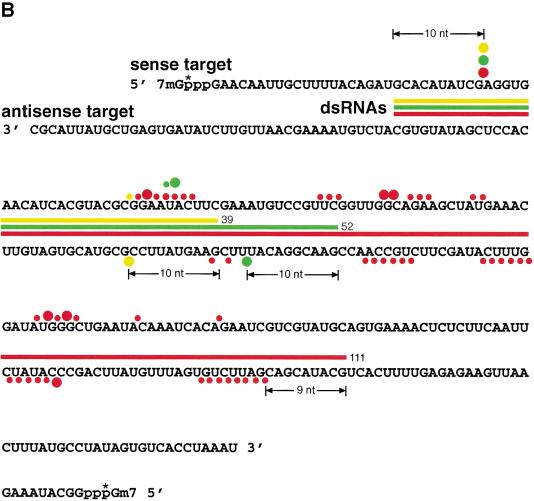
Figure 3.
Mapping of sense and antisense target RNA cleavage sites. (A) Denaturing gel electrophoresis of the stable 5′ cleavage products produced by 1 h incubation of 10 nM sense or antisense RNA 32P-labeled at the cap with 10 nM dsRNAs of the p133 series in Drosophila lysate. Length markers were generated by partial nuclease T1 digestion and partial alkaline hydrolysis (OH) of the cap-labeled target RNA. The regions targeted by the dsRNAs are indicated as black bars on both sides. The 20–23-nt spacing between the predominant cleavage sites for the 111-bp dsRNA is shown. The horizontal arrow indicates unspecific cleavage not caused by RNAi. (B) Position of the cleavage sites on sense and antisense target RNAs. The sequences of the capped 177-nt sense and 180-nt antisense target RNAs are represented in antiparallel orientation such that complementary sequence are opposing each other. The region targeted by the different dsRNAs are indicate by differently colored bars positioned between sense and antisense target sequences. Cleavage sites are indicated by circles (large circle for strong cleavage, small circle for weak cleavage). The 32P-radiolabeled phosphate group is marked by an asterisk.