Figure 6.
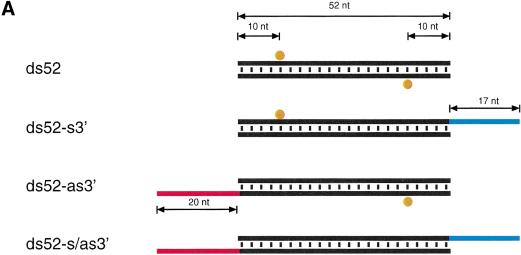
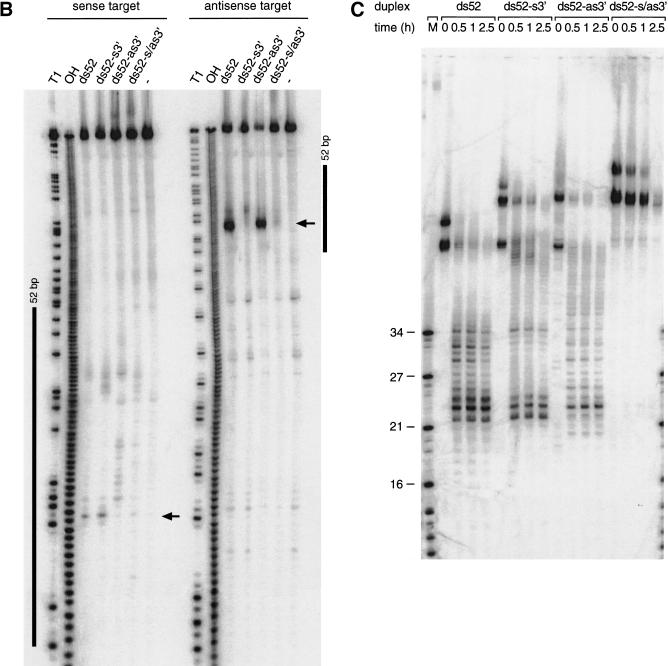
Long 3′ overhangs on short dsRNAs inhibit RNAi. (A) Graphic representation of 52-bp dsRNA constructs. The 3′ extensions of sense and antisense strand are indicated in blue and red, respectively. The observed cleavage sites on the target RNAs are represented as orange circles analogous to Figure 4A and were determined as shown in B. (B) Position of the cleavage sites on sense and antisense target RNAs. The target RNA sequences are as described in Figure 3B. DsRNA (10 nM) was incubated with target RNA at 25°C for 2.5 h in Drosophila lysate. The stable 5′ cleavage products were resolved on the gel. The major cleavage sites are indicated with a horizontal arrow and are also represented in A. The region targeted by the 52-bp dsRNA is represented as a black bar at both sides of the gel. (C) Processing of 52-bp dsRNAs with different 3′ extensions. Internally 32P-labeled dsRNAs (5 nM) were incubated in Drosophila lysate, and reaction aliquots were analyzed at the indicated time points. An RNA size marker (M) has been loaded in the left lane, and the fragment sizes are indicated. Double bands at time zero are caused by incompletely denatured dsRNA.