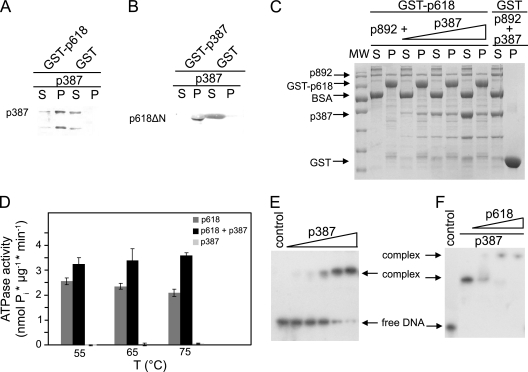
Fig. 5.
p387 protein-protein interactions. (A) Coprecipitation experiments were carried out with p387 and GST-p618. (B) The binding site of p387 on p618 was mapped using a p618 fragment lacking the N-terminal domain (p618ΔN). Assays were performed with 0.2 mg/ml of target proteins, and GST conjugated to GSH beads and BSA served as controls. (C) A competition pulldown experiment with p618 and p892 and with increasing concentrations of p387 (0.1 to 0.3 mg/ml) was performed as described above. Supernatant (S) and pellet (P) fractions were analyzed either by SDS-PAGE with Coomassie staining or by Western blotting (A and B). A molecular size standard (MW) was used to estimate the apparent molecular protein masses. (D) The effect of a 5-fold molar excess of p387 on the ATPase activity of p618 was estimated at 650 nm at the indicated temperatures in the presence of 1 mM ATP. (E and F) Electrophoretic mobility shift assays. (E) DNA binding of p387 was investigated at 50°C for 25 min with 1.5 nM DNA (147 bp) and increasing concentrations of protein (0 to 5 μM p387). (F) Binding of p618 to the DNA-p387 complex (molar ratio of p618 to p387, 0.5 to 2) resulted in the formation of a multimolecular complex. The negative controls contained substrate DNA only. Protein-bound and free DNAs were separated by electrophoresis on an 11% nondenaturing acrylamide gel and were visualized by autoradiography.