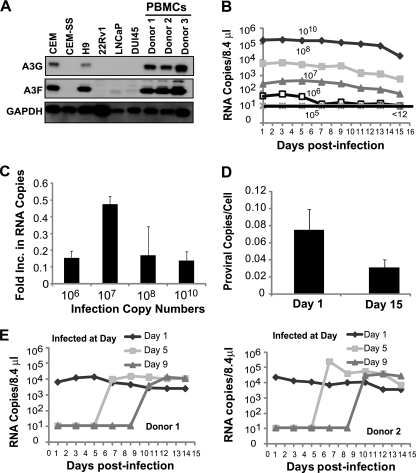
Fig. 3.
Replication of XMRV in PBMCs. (A) A3G and A3F expression in T cell lines (CEM, CEM-SS, and H9), prostate cancer cell lines (22Rv1, LNCaP, and DU145), and PBMCs from three different donors. Western blotting employed an equal amount of protein (10 μg); A3G or A3F protein expression was detected using anti-A3G (C-17) or anti-A3F (C-18) antibodies. GAPDH served as a loading control. (B) PBMCs (1 × 107 cells) from donor 2 were infected with different amounts of XMRV as described for Fig. 1A, and culture supernatants were harvested every 2 days. XMRV RNA copies were determined as described for Fig. 1A. Results are plotted as XMRV RNA copy number per 8.4 μl of virus supernatant. A representative experiment of three independent experiments from three different donors is shown. (C) Increase in viral production in PBMCs. The increase in viral production was determined by dividing the average RNA copy number from days 13 to 15 by the average RNA copy number from days 1 to 3. The averages of three independent experiments using three different donors are shown. The error bars indicate standard errors of the means. (D) Proviral copy numbers per cell in infected PBMCs from three donors. Infected PBMCs were harvested at day 1 and day 15, genomic DNA was isolated, and equal amounts of DNA were used to detect XMRV gag and CCR5 by quantitative real-time PCR. Error bars represent the standard errors of the means. (E) Infection of PBMCs (1 × 107 cells) from donor 1 (left) and donor 2 (right) on days 1, 5, and 9 after activation. PBMCs were infected on the specified days postactivation with XMRV (108 RNA copies) harvested from 22Rv1 cells. Culture supernatants were harvested every second day, and XMRV RNA was detected by quantitative real-time RT-PCR. The results are plotted as copies of viral XMRV RNA per 8.4 μl of culture supernatant of infected cells.