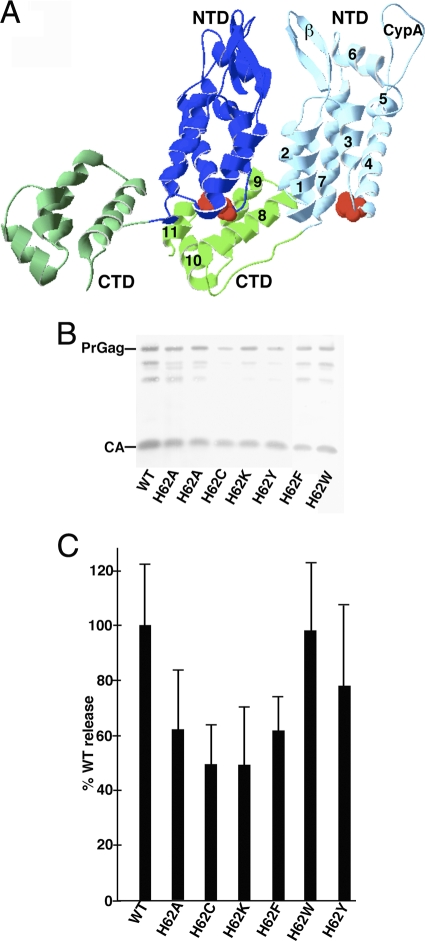
Fig. 1.
Effects of mutations on virus particle assembly and release. (A) Shown is a pair of CA proteins (Protein Data Bank [PDB] accession no. 3GV2) in HIV-1 CA hexamers viewed roughly from the center of the hexamer outward. NTDs (blue) and CTDs (green) are marked, as are the β-hairpin (β) and cyclophilin A binding (CypA) loops and helices 1 to 11 on one of the capsid proteins. H62 residues are indicated in red. (B) HEK 293T cells were transfected with the indicated HIVLuc constructs. At 72 h posttransfection, virus samples were collected, and Gag proteins were detected after gel electrophoresis by immunoblotting using a primary anti-CA antibody. Full-length PrGag and CA bands are as indicated. (C) For quantification of viral particle release at 72 h posttransfection, virus and cell samples were collected and subjected to electrophoresis and immunoblotting using an anti-CA antibody. Cell and virus Gag levels were quantitated densitometrically; raw virus release levels (Virus Gag/Cell PrGag) were calculated and are shown, normalized to WT HIV release levels. Results derive from two (H62K and H62W), three (H62C and H62F), four (H62Y), five (H62A), or eight (WT) separate experiments. Standard deviations are as shown.