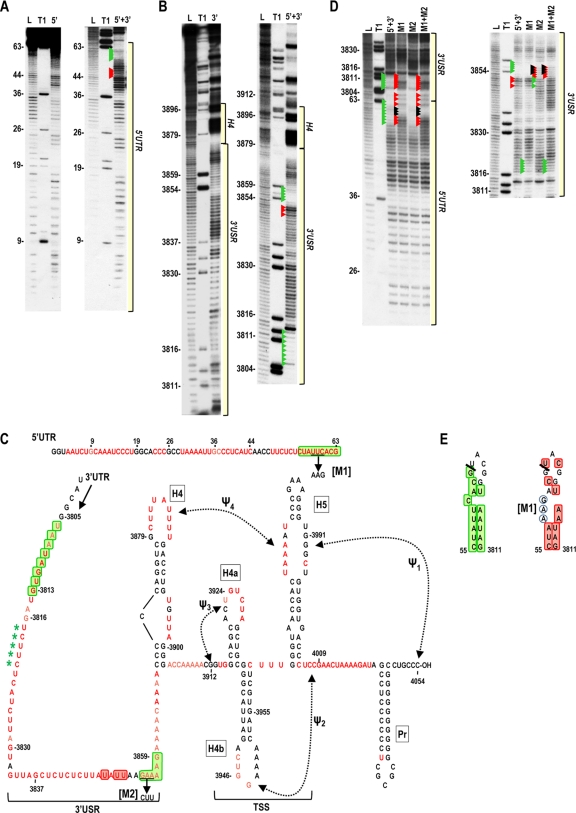
Fig. 7.
In-line cleavage profile of covalently linked 5′ UTR and 3′ UTR. Transcripts containing the 5′ UTR covalently linked to the 3′ UTR (5′+3′) were 5′ end labeled and subjected to in-line cleavage. (A) Comparison of 5′+3′ and 5′ UTR (5′) cleavage profiles. Residues with enhanced or reduced cleavage in 5′+3′ compared with the 5′ UTR alone are denoted by red and green arrowheads, respectively. The location of the 5′ UTR region within the 5′+3′ cleavage profile is shown. See the legend to Fig. 5 for other abbreviations. (B) Comparison of the 5′+3′ and 3′ UTR (3′) cleavage profiles. Residues with enhanced or reduced cleavage in 5′+3′ compared with the 3′ UTR alone are denoted by red and green arrowheads, respectively. The locations of 3′ USR and 3′ hairpins are indicated. Longer electrophoresis of samples did not reveal any additional changes downstream of this region (not shown). (C) Locations of residues with enhanced (red boxes) or reduced (green boxes) flexibility in 5′+3′ compared with the individual UTR fragments. See the legend to Fig. 5 for more details. Mutations (M1 and M2) generated to access a possible interaction between two sequences that had reduced flexibility in 5′+3′ are shown. Asterisks denote loss of cleavage at these positions in the M1 mutant 5′+3′ fragment. (D) In-line cleavage profile for 5′+3′ transcripts containing either M1, M2, or both mutations (M1+M2). Arrowheads in the 5′+3′ lane denote differences from the profiles of the individual fragments (from panels A and B). Red and green arrowheads in the mutant cleavage lanes denote differences from the wt 5′+3′ transcripts. Black arrowheads denote bases that were mutated. (E) Possible interaction between the 5′ and 3′ UTR regions in 5′+3′. Left, green boxes denote residues with reduced flexibility in the 5′+3′ fragment compared with the individual UTRs. The thick line denotes the junction between the 5′ and 3′ UTRs in the 5′+3′ fragment. Right, red boxes denote enhanced flexibility of residues in 5′+3′ transcripts containing M1 mutations (circled). Note that the altered sequence remains inflexible (5′AAGA) and can now pair with sequence in the 3′ UTR region of the 5′+3′ fragment (5′UCUU) that also showed reduced flexibility in the m1 fragment.