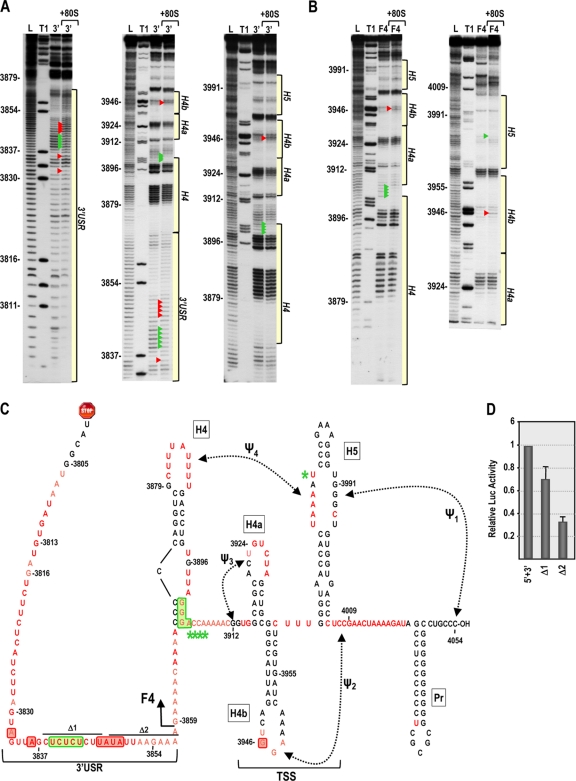
Fig. 8.
In-line cleavage profiles of the 3′ UTR and fragment F4 in the presence and absence of 80S ribosomes. (A and B) In-line cleavage autoradiograph of labeled 3′ UTR (A) or fragment F4 (B) in the presence (+80S) and absence of 80S ribosomes. Green and red arrowheads denote residues whose cleavage pattern was altered by the presence of ribosomes in all three replicates. See the Fig. 5 legend for more details. (C) Structure of the 3′ UTR of TCV. Residues in red are flexible and susceptible to cleavage, with color intensity reflecting the degree of cleavage. Green and red boxes denote residues in the 3′ UTR fragment that have reduced or enhanced susceptibility to cleavage, respectively, in the presence of 80S salt-washed ribosomes. F4 shared the 80S-induced enhanced cleavage at position 3946 only, and asterisks denote residues in F4 with reduced flexibility in the presence of 80S ribosomes. Locations of deletions Δ1 and Δ2, used to assay for effect on translation, are shown. (D) Residues with altered cleavages in the 3′ USR upon ribosome binding are important for translation. The reporter construct containing the TCV 5′ UTR and 3′ UTR+ was engineered to delete the segments shown in panel C. RNA transcripts were assayed for translation in protoplasts. Standard deviations from three independent experiments are shown.