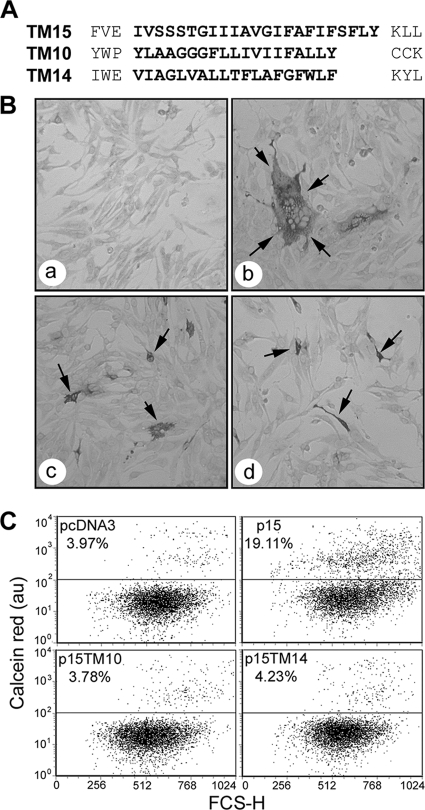
Fig. 1.
The FAST protein TMDs do not function modularly within the context of p15. (A) Linear representation of the TMD regions of p15, ARV p10, and p14. Sequences in bold delineate the predicted TMDs and the sequences that were exchanged to create chimeric p15 proteins whose TMDs were replaced by those of p14 (TM14) or p10 (TM10). (B) QM5 cells were transfected with pcDNA3 (a), authentic p15 (b), p15TM10 (c), or p15TM14 (d) and immunostained using polyclonal anti-p15 antiserum at 9 (a and b) or 24 (c and d) h posttransfection. Arrows indicate antigen-positive syncytia (b) or single-cell foci (c and d). (C) Dual-color pore formation assay. QM5 cells were cotransfected with pEGFP and pcDNA3, authentic p15, p15TM10, or p15TM14 and overlaid with calcein red-labeled Vero cells. Cells were resuspended and fixed at 9 h posttransfection, and EGFP-positive cells were gated. The dot plots are representatives of duplicate samples from a single experiment, and the average percentage of EGFP-expressing cells that acquired calcein red due to pore formation is shown above the horizontal gating line, relative to forward scatter (FSC-H). The indicated percentage values varied by <1% between the duplicate samples and between two separate experiments.