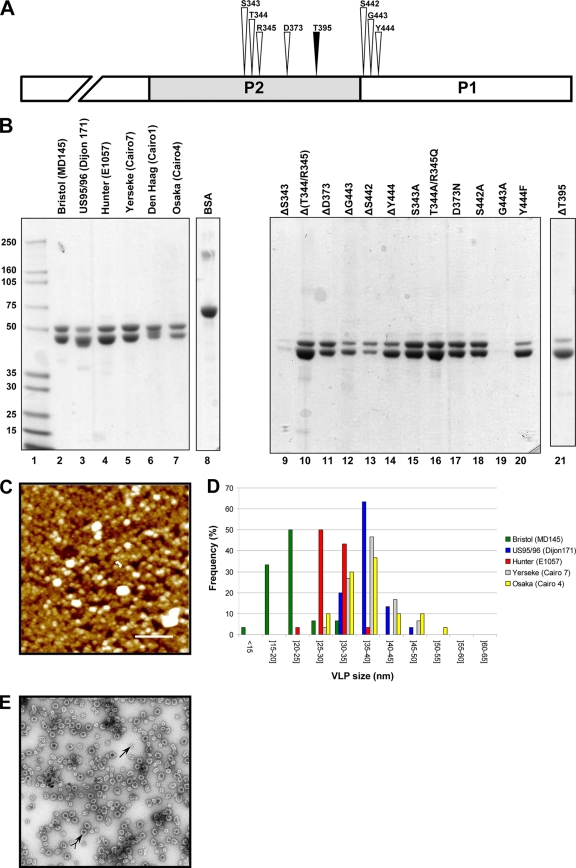
Fig. 1.
Characterization of the purified VLPs of the GII.4 variants. (A) Drawing to scale of the amino acid residues (S343, T344, R345, D373, S442, G443, and Y444) from the α-fucose binding site (white arrowhead) (8, 49) and the inserted threonine residue, T395 (black arrowhead). The NH2 terminus of the P1 domain is truncated. (B) SDS-PAGE analysis of the VLPs from the GII.4 variants (lanes 2 to 7) and the mutagenized Hunter VLPs (lanes 9 through 21). The name of the NoV strain is indicated in parentheses for each variant. The mutagenized VLPs originated from the Hunter variant (strain E1057), and the location of the amino acid residues is based upon the ORF2 amino acid sequence of the E1057 strain (GenBank accession number EU876890). Two micrograms of bovine serum albumin (Pierce) was added as a control for the protein estimation (lane 8). Lane 1 contains the protein molecular size markers that are indicated on the right side of the gel. (C) Atomic force microscopy imaging of the Hunter purified VLPs. The aggregates are shown in white. One of the VLPs is indicated by a bracket. Scale bar, 250 nm. (D) Size distribution of the VLPs as determined from atomic force microscopy imaging. The variant VLPs are color coded according to the legend on the right side of the graph. (E) Electron micrograph of the CsCl-purified Hunter VLPs after negative staining. Complete and subunit VLPs are indicated by arrows with and without a tail, respectively.