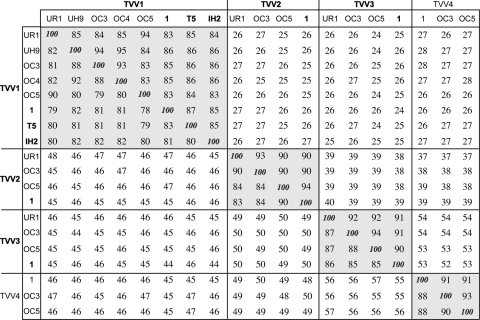
Fig. 2.
Pairwise sequence comparisons of the 19 TVV sequences. Percent identities for pairwise comparisons of the full-length genomic nucleotide sequences are shown below and left of the 100% diagonal, and those of the predicted CP/RdRp amino acid sequences are shown above and right of that diagonal. Pairwise alignments were performed by the program EMBOSS::needle(global), with default settings, at http://www.ebi.ac.uk/Tools/emboss/align. Clusters of percent identity scores of ≥78%, reflecting the four trichomonasvirus species (TVV1, -2, -3, and -4), are highlighted by background shading. Previously reported species and strains are indicated by boldface.