Abstract
The establishment of expression domains of developmentally regulated genes depends on cues provided by different concentrations of transcriptional activators and repressors. Here we analyze the regulation of the Drosophila gene zen, which is a target of the Decapentaplegic (Dpp) signaling pathway during cellular blastoderm formation. We show that low levels of the Dpp signal transducer p-Mad (phosphorylated Mad), together with the recently discovered negative regulator Brinker (Brk), define the spatial limits of zen transcription in a broad dorsal-on/ventral-off domain. The subsequent refinement of this pattern to the dorsal-most cells, however, correlates with high levels of p-Mad that accumulate in the same region during late blastoderm. Examination of the zen regulatory sequences revealed the presence of multiple Mad and Brk binding sites, and our results indicate that a full occupancy of the Mad sites due to high concentrations of nuclear Mad is the primary mechanism for refinement of zen. Interestingly, several Mad and Brk binding sites overlap, and we show that Mad and Brk cannot bind simultaneously to such sites. We propose a model whereby competition between Mad and Brk determines spatially restricted domains of expression of Dpp target genes.
Keywords: Dpp morphogen, target genes, Smad activation, Brk repression
The TGF-β superfamily of secreted signaling molecules represents a group of evolutionarily-conserved proteins that control multiple cellular processes in a range of organisms (for reviews, see Massagué 1998; Massagué et al. 2000). The cellular responses to TGF-β ligands are mediated by a highly conserved signal transduction pathway involving a family of transmembrane receptor serine/threonine kinases and cytoplasmic signal transducers, the Smad proteins. The activated receptors phosphorylate the receptor-regulated Smads that form complexes with the co-Smads, translocate to the nucleus, and regulate the expression of target genes by direct interaction with DNA or other transcription factors (for reviews, see Massagué and Wotton 2000; ten Dijke 2000).
There are several TGF-β family members in Drosophila, among which Dpp is the best-studied (for review, see Podos and Ferguson 1999). The Dpp signal is transduced to the nucleus by Smad complexes containing the receptor-regulated Mad protein and the co-Smad Medea (for review, see Raftery and Sutherland 1999). Mad and Medea have been shown to bind DNA and activate several Dpp target genes. For example, Mad and Medea bind to specific sites in the Dpp response element of the tinman (tin) gene, the tin-D enhancer (Xu et al. 1998), and these sites were shown to be essential for normal tin expression in the embryonic visceral mesoderm. Direct Mad–DNA contact also plays a role in the transcriptional activation of the Ubx gene in the developing midgut (Eresh et al. 1997) and the vestigial (vg) gene in the imaginal wing disc (Kim et al. 1996, 1997).
Mad/Medea binding sites contain repeats of the degenerate sequence GNCN, which is consistent with the sequence of the Smad binding element (SBE) GTCT found in the response regions of TGF-β and activin target genes (for review, see ten Dijke 2000). However, the low complexity of the recognition sites and their low affinity for Smad binding (Shi et al. 1998) cannot explain the highly specific target gene responses to TGF-β signaling. It was therefore proposed that in many cases Smad proteins achieve specific interactions with cognate DNA by interacting with DNA-binding partners (for review, see ten Dijke 2000).
One interesting feature of Dpp and other members of the TGF-β family, such as activin and the bone morphogenic proteins (BMPs), is that they can function as morphogens (for review, see Podos and Ferguson 1999). Morphogens induce different cell fates at different concentrations or activities (for review, see Lawrence and Struhl 1996). In both Drosophila and Xenopus embryos, gradients of Dpp/BMP activity are established that are responsible for patterning along the dorsoventral axis (Ferguson and Anderson 1992; Wharton et al. 1993; Wilson et al. 1997). Dpp activity has its highest levels along the dorsal midline of the cellular blastoderm embryo and declines toward more lateral regions where it is inhibited by the product of the short gastrulation (sog) gene (Holley et al. 1995; Biehs et al. 1996; Marqués et al. 1997). The high levels determine the cell fate of the amnioserosa in the dorsal-most cells, whereas lower levels specify aspects of the dorsal epidermis in dorsolateral cells. The absence of Dpp activity in ventrolateral regions permits the formation of the neurogenic ectoderm, which gives rise to both the ventral epidermis and the central nervous system.
How does Dpp specify cell fate in a concentration-dependent manner? It is thought that Dpp signaling in the early embryo regulates the transcription of downstream target genes that are expressed in nested domains centered around the dorsal midline (Jazwinska et al. 1999a,b; Ashe et al. 2000). High-level Dpp targets such as Race (Ashe and Levine 1999) and u-shaped (ush; Cubbada et al. 1997) are expressed in the presumptive amnioserosa. pannier (pnr; Winick et al. 1993) is expressed in a broader domain that spans the amnioserosa and part of the dorsal ectoderm. Thus, it requires lower levels of Dpp. Finally, low-level targets such as early zen (Doyle et al. 1986) and dpp (St. Johnson and Gelbart 1987) are expressed in an even broader domain that abuts the ventral ectoderm. A possible molecular mechanism to explain the threshold responses of Dpp target genes is that their promoters have different affinities to Smads and therefore can be induced by different levels of nuclear Smads, similar to the mechanism of differential activation by the Drosophila morphogens Dorsal (Dl; Jiang and Levine 1993) and Bicoid (Bcd; Driever et al. 1989; Simpson-Brose et al. 1994). That an additional mechanism is involved came from the characterization of the brinker (brk) gene (Campbell and Tomlinson 1999; Jazwinska et al. 1999a,b; Minami et al. 1999). brk negatively regulates low-level and intermediate-level target genes. Study of the response elements of these target genes can therefore provide clues about the mechanisms of threshold responses to the Dpp morphogen, as well as the interplay of positive and negative inputs in the expression of target genes.
We have focused our studies on the target gene zen, which has a dynamic pattern of expression in the early embryo. During precellular nuclear division cycles 11–13 and during early cellularization (nuclear cycle 14), zen is expressed in a broad dorsal-on/ventral-off pattern. This pattern is thought to be activated by an unknown ubiquitous activator present throughout the embryo and repressed by the Dl morphogen localized in ventral regions (Rushlow et al. 1987). It is Dpp-independent because early zen expression is normal in dpp null mutants (Rushlow and Levine 1990; Ray et al. 1991). However, slightly later, during early to mid-cellularization, maintenance of the zen pattern becomes dependent on Dpp because zen transcripts fade away suddenly in dpp null mutants (Rushlow and Levine 1990; Ray et al. 1991). It also becomes dependent on Brk repression because zen transcripts expand into the ventral ectoderm in brk mutants (Jazwinska et al. 1999b). Thus, the broad pattern of zen is maintained by Dpp in the dorsal region and repressed by Brk in ventral regions. During mid- to late cellularization, this pattern undergoes a process of refinement in which zen transcripts are lost from the lateral regions and become restricted to a narrow domain of the dorsal-most cells. Brk plays no role in refinement because in brk mutants, although zen expands ventrally, it refines normally (Jazwinska et al. 1999b).
zen expression is directed by 1.6 kb of 5′ flanking DNA sequences referred to as the zen promoter (Doyle et al. 1989). The distal part of the promoter between −1.2 and −1.4 kb is responsible for Dl-dependent ventral repression (Jiang et al. 1993; Kirov et al. 1993). Sequences required for the initiation, maintenance, and refined expression of zen are located in the proximal 0.7 kb of the promoter, but they are not well-characterized (Doyle et al. 1989).
Here we address the question of how the Dpp pathway components Mad and Medea, and the negative regulator Brk, mediate maintenance and refinement of zen transcription. We find that lowering the level of Dpp signaling does not perturb maintenance but abolishes refined expression of zen. In addition, immunostaining with antibodies that recognize activated Mad proteins show that high levels of activated Mad are required for refinement, whereas lower levels are sufficient for maintenance. Our molecular analysis shows that Mad, Medea, and Brk regulate zen transcription by binding to specific and partially overlapping sites on the zen promoter. We propose that a simple competitive mechanism might be involved in the transcriptional regulation of zen and possibly other Dpp target genes.
Results
zen maintenance requires less Dpp signaling than zen refinement
To better understand the role of the Dpp activity gradient in zen regulation, we examined the expression pattern of zen in embryos carrying dpp hypomorphic alleles (Wharton et al. 1993). dpp alleles can be ordered in an allelic series of increasing strength as measured by two phenotypes: percentage of dominant lethality and degree of ventralization of the embryonic cuticle. Null alleles are haploinsufficient, showing more than 95% lethality when heterozygous. Embryos homozygous for null alleles show a complete loss of all dorsal ectodermal structures and expansion of the ventral denticle belts around the entire circumference of the embryo. The hypomorphic alleles display a range of both phenotypes. We analyzed the zen expression pattern in embryos carrying the hypomorphic alleles dpphr4, dpphr27, and dppH94. dpphr4 is the weakest allele, showing about 5% dominant lethality. dpphr27 and dppH94 show 30%–40% and 70%–80% dominant lethality, respectively. Both dpphr4and dpphr27 homozygous embryos show complete loss of amnioserosa, with dpphr27 showing an additional loss of dorsal ectodermal structures as well as the expansion of the ventral denticle belts. In homozygous dppH94 embryos, almost all dorsal structures are lost and the ventral denticle belts expand even further toward the dorsal midline.
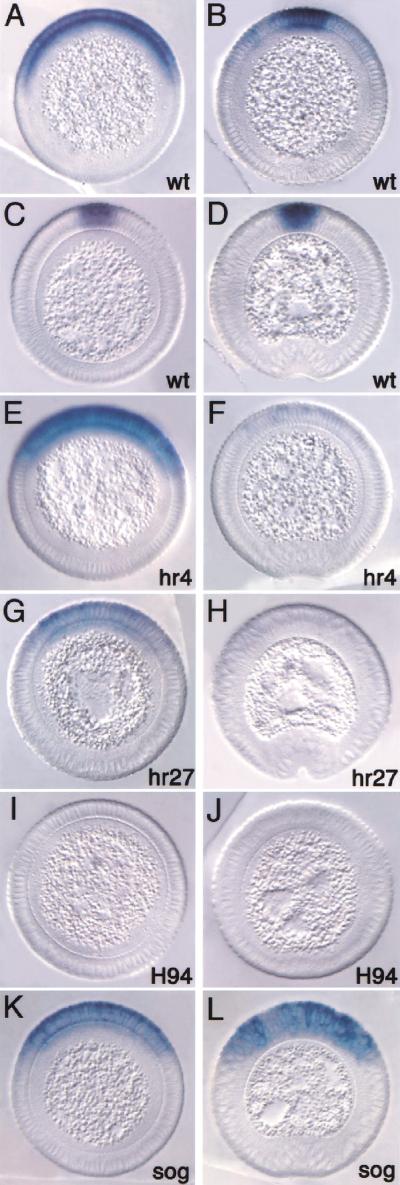
The early broad zen pattern seen in precellular stages and during early cellularization (Fig. 1A) is normal in dpphr4, dpphr27, and dppH94 homozygous embryos (data not shown). During mid- to late cellularization, zen expression in dpphr4 homozygotes was maintained (Fig. 1E), whereas a reduction in the level of zen RNA was observed in dpphr27 homozygotes (Fig. 1G). zen expression was completely lost in the stronger hypomorphic dppH94 homozygotes (Fig. 1I), similar to that seen in dpp null embryos (Rushlow and Levine 1990; Ray et al. 1991). This indicates that a certain level of Dpp activity, still present in dpphr4 but progressively depleted in the stronger mutants, is required to maintain zen expression in its broad domain. Importantly, during mid- to late cellularization, zen transcripts did not refine in dpphr4 and dpphr27 homozygotes (Fig. 1E,G) and diminished rapidly by gastrulation (Fig. 1F,H). This is in striking contrast to wild-type embryos in which zen refines during mid- to late cellularization so that transcripts become restricted to the presumptive amnioserosa (Fig. 1B,C) and persist through gastrulation (Fig. 1D). The above results indicate that the level of Dpp signaling plays an important role in the dynamic changes in zen expression during cellularization. The refined zen expression is especially sensitive to the level of Dpp activity because it is abolished in the weak hypomorphic mutants.
Figure 1.
Dpp-dependent regulation of zen expression. Cross sections of embryos hybridized with zen antisense RNA probes. Dorsal is up. (A–D) Wild type, (E,F) dpphr4/dpphr4, (G,H) dpphr27/dpphr27, (I,J) dppH94/dppH94, (K,L) sogYS06/Y. (A) Embryo in early to mid-cellularization showing the broad dorsal-on/ventral-off zen pattern. (B) Embryo in mid-cellularization undergoing zen refinement. (C) Embryo in mid- to late cellularization showing the refined zen pattern restricted to the presumptive amnioserosa. (D) Embryo undergoing gastrulation with continued strong zen expression. Remaining embryos on the left are in mid- to late cellularization and can be compared with (C), and those on the right are beginning gastrulation and can be compared with (D). Note the decrease in the level of zen transcripts during mid- to late cellularization with decreasing dpp activity, and the loss of expression by gastrulation. Although a reduction in the level of transcripts was observed in dpphr27 embryos, the ventral limit of zen expression did not change significantly. In the absence of sog, zen expression is maintained to gastrulation, but there is no refinement.
High levels of activated Mad drive zen refined expression
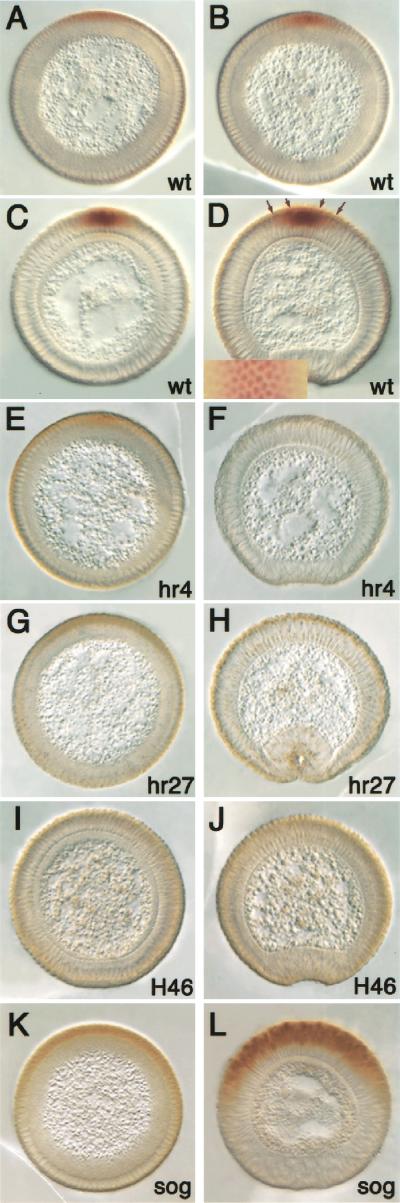
Recently, it was shown that the level of Dpp activity in imaginal discs and embryos can be correlated with the amount of nuclear Smads (Sutherland and Raftery 2000; Tanimoto et al. 2000). We used the same antibodies raised against the mammalian phosphorylated Smad1 (Persson et al. 1998), which also recognize Drosophila phosphorylated Mad (p-Mad; Tanimoto et al. 2000), to determine if the p-Mad staining pattern can be correlated with zen transcriptional activity in wild-type and dpp hypomorphic mutant embryos. p-Mad staining in wild-type embryos was first detected during early to mid-cellularization in a dorsal domain spanning about 20% of the embryonic circumference (Fig. 2A). As cellularization proceeds, staining becomes stronger in the dorsal-most cells and decreases in the dorsolateral region (Fig. 2B,C). By gastrulation, staining is mostly localized to the presumptive amnioserosa (five to six cells; Fig. 2D, inner arrows) with weaker staining observed in the dorsolateral regions (three to four cells to either side; Fig. 2D, outer arrows). Staining is nuclear and gradually dissipates laterally (Fig. 2D, inset). The strongest p-Mad staining correlates exactly with refined zen expression (cf. Figs. 1C,D and 2C,D), indicating that high levels of p-Mad are required for zen transcription in the refined domain. Interestingly, the process of p-Mad accumulation is similar to that of zen refinement in that a broad pattern becomes restricted into a narrow pattern (cf. Figs. 1A–D and 2A–D). Hence, the refinement of zen is a reflection of the formation of the Dpp activity gradient.
Figure 2.
High levels of activated Mad correlate with refined zen expression during late cellularization. Cross sections of embryos stained with anti-phospho-Smad antibodies. Dorsal is up; cellularization stages are as in Fig. 1. (A–D) Wild type, (E,F) dpphr4/dpphr4, (G,H) dpphr27/dpphr27, (I,J) dppH94/dppH46, (K,L) sogYS06/Y. (A–D) p-Mad accumulates as cellularization proceeds, reaching highest levels in the presumptive amnioserosa. The inset in D shows a surface view of a region of the dorsal side of a whole-mount staining, the same region delimited by the outer arrows on the section. Note that staining gradually decreases laterally. (E–H) p-Mad accumulation to high levels is not observed in dpp mutants. (I,J) In dppH46 embryos, no p-Mad staining is observed, indicating that the antibody is specific for Mad. (K,L) In sog mutants, p-Mad accumulates in a broad domain but may not reach the peak levels seen in the wild type.
We further analyzed p-Mad staining in dpp and sog mutant embryos. The hypomorphic dpphr4 and dpphr27 embryos showed weak p-Mad staining at mid-cellularization (Fig. 2E,G, respectively) that disappeared by gastrulation (Fig. 2F,H), whereas the amorphic dppH46 embryos showed no p-Mad staining at any stage (Fig. 2I,J). Similarly, during mid-cellularization zen transcripts are lost prematurely in the strong dpp mutant, but not in the weaker mutants (Fig. 1E,G,I).
One can therefore conclude that the residual p-Mad signaling in the dpp hypomorphs is sufficient for zen maintenance. However, high levels of p-Mad are required for refined zen expression because both p-Mad staining and zen transcripts were not detected during late cellularization even in the weakest mutant (Fig. 1F,H,J). In sog mutants, the high-level p-Mad domain is broadened, as is zen transcription (cf. Figs. 1L and 2L).
Smad and Brk binding to zen promoter DNA is essential for proper zen expression
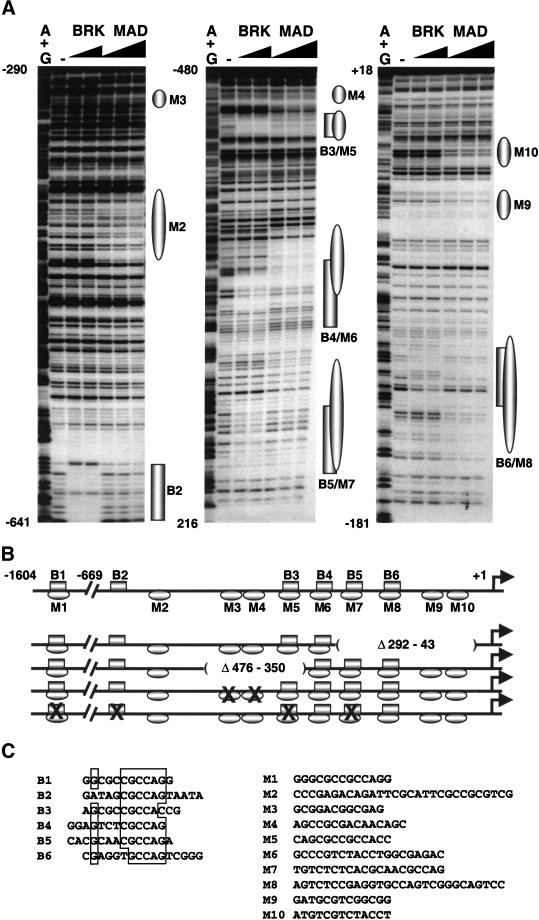
Biochemical and genetic evidence indicate that the DNA-binding activity of Drosophila Smad proteins is essential for the expression of Dpp target genes (Kim et al. 1997; Xu et al. 1998). Because our results implied that p-Mad is involved in the activation of zen expression, we screened the 1.6-kb zen regulatory region (zen promoter) for Mad and Medea DNA binding sites by EMSA (gel-shift) and DNase I footprinting assays (see Materials and Methods). Mad specifically bound to 10 sites (M1–M10), 9 of which are located in the proximal region of the zen promoter between positions −571 bp and −124 bp upstream of the zen transcription start site (Fig. 3A,B). The co-Smad protein, Medea, was tested by gel shift with DNA fragments and oligonucleotides spanning the promoter and showed the same binding pattern as Mad (data not shown). The Mad/Medea binding sites are G/C rich, similar to the Mad sites on the vg enhancer and the Mad/Medea sites on the tin-D enhancer (Kim et al. 1997; Xu et al. 1998). All sites contain the tandemly repeated sequence GNCN, which is present in Smad response elements of Dpp and TGF-β/activin-inducible genes (for review, see Raftery and Sutherland 2000; ten Dijke 2000).
Figure 3.
Mad and Brk bind to the zen promoter. (A) DNase I footprinting analysis of Mad and Brk GST fusion proteins bound to the proximal 669 bp of the zen promoter. Increasing amounts of Brk (50 ng and 150 ng) and Mad (500 ng, 1500 ng, and 4500 ng) were incubated with zen-promoter DNA fragments: (left panel) EcoRI–AccI fragment labeled at the EcoRI site (−669 to −290), (middle panel) XbaI–AvaI fragment labeled at the XbaI site (−198 to −480), (right panel) XbaI–BamHI fragment labeled at the XbaI site (−198 to +18). The numbers correspond to the positions of the nucleotides relative to the transcription start site (+1). The (−) lanes show DNase I digestion of the DNA probes. The G+A lanes show the chemical degradation of the probes on G+As. Regions protected by Mad and Brk proteins are depicted as ovals and rectangles, respectively. (B) Schematic representation of the Smad (Mad/Medea) and Brk binding sites on the 1604-bp zen promoter, and the deletion and point mutations used in the transgenic analysis. The drawing is in scale only for the proximal 669 bp of the promoter. The locations of Brk binding sites (B2–B6) and Mad sites (M2–M10) are based on the footprinting data. The location of B1/M1 and all of the Medea sites were found by gel-shift analysis (not shown). The deletions are designated as absent lines inside parentheses, and the binding sites with point mutations are designated by X. (C) Alignment of the sequences of the BRK binding sites (B1–B6) (left). A list of Mad/Medea sites that cannot be aligned because of their degeneracy (right). Note that none of the Mad/Medea sites contains the inverted repeat 5′-GTC TAGAC-3′, which was determined as an optimal site for Smad3 and Smad4 proteins (Zawel et al. 1998).
Sites M2, M6, M7, and M8 contain the “Smad box” GTCT motif, which can bind a single MH1 domain (Shi et al. 1998). The sites have different affinities for the Mad protein as seen in the extent of DNA protection (compare footprints in a lane in Fig. 3A).
As mentioned earlier, zen is repressed in the ventral ectoderm by Brk during mid-cellularization. The possibility that Brk might bind DNA was predicted based on the HTH fold in the N-terminal region of the Brk protein and a weak homology of this same region with the homeodomain (Campbell and Tomlinson 1999; Jazwinska et al. 1999a). DNA-binding experiments performed with DNA from the zen promoter using recombinant GST–Brk fusion proteins confirmed our prediction. There are six Brk binding sites on the zen promoter (B1–B6), five of which are located in the proximal part of the promoter (Fig. 3A,B). An alignment of the sites shows that four of them contain the sequence 5′-CGCCAG-3′, and the other two have a single base change of this canonical sequence (Fig. 3C). By comparing the degree of protection from DNase I when using a particular amount of protein (Fig. 3A, cf. footprints within a lane), it appears that the Brk binding sites differ in their affinities for the recombinant Brk protein by two- to fivefold (Fig. 3A).
Interestingly, the DNase I footprints of Brk overlap extensively with the sites protected by Mad and Medea (Fig. 3A,B), with the exceptions of sites B2, M2, M3, M4, M9, and M10.
We tested the functional relevance of the binding sites in vivo by using zen-lacZ transgenes. Previous transgenic experiments showed that a truncated zen promoter containing only the proximal 293 bp cannot drive any expression of the lacZ reporter in blastoderm embryos. However, a promoter containing the proximal 670 bp can drive strong dorsal expression and refinement, although there is no ventral repression by Dl (Doyle et al. 1989). To further delineate sequences involved in the maintenance and refinement of zen expression, we tested several deletions within the proximal region in the context of the full-length 1.6-kb zen promoter (schematized in Fig. 3B). Deletion Δ292–43, which eliminates two Mad/Medea sites (M9,M10; see Fig. 3B) and two Brk/Mad/Medea sites (B5M7, B6M8), drives normal expression during early cellularization but fails to refine later (Fig. 4, cf. A and B). Similarly, the smaller deletion Δ476–350, which eliminates M3, M4, and B3M5, failed to undergo refinement (Fig. 4C). In deletion Δ161–131, only the B6M8 site is missing, and 70%–80% of the transgenic embryos show refinement (data not shown). The deletion Δ217–178, which does not eliminate any sites, has no effect on the expression pattern of lacZ (data not shown). To determine if the lack of refinement observed in the large deletions might be due to the loss of Smad sites, the two Mad/Medea sites located between −476 and −350 were mutated (M3 and M4; see Fig. 3B). Embryos carrying this double mutation showed a lack of refinement (Fig. 4D), similar to the deletion Δ476–350. These results indicate that zen refinement requires all of the Smad sites in the proximal promoter.
Figure 4.
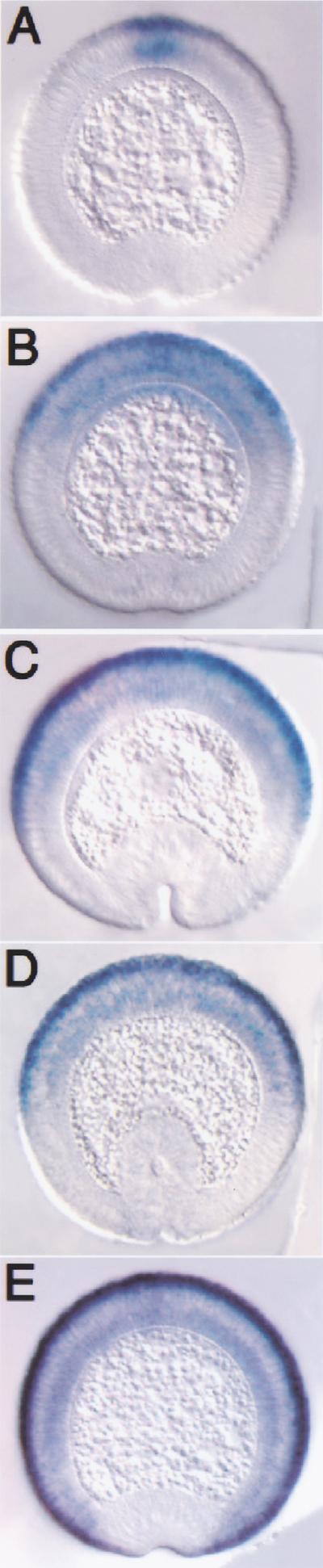
Mutation of Smad and Brk binding sites results in altered expression of a zen–lacZ reporter. Cross sections of transgenic embryos undergoing gastrulation carrying the zen–lacZ fusion constructs (described in Fig. 3B) hybridized with lacZ antisense RNA probes. Dorsal is up. (A) Embryo carrying the full-length 1.6-kb zen promoter-lacZ shows refinement. (B) Embryo carrying the deletion Δ292–43. (C) Embryo carrying the deletion Δ476–350. (D) Embryo carrying a mutation that eliminates Smad binding sites M3 and M4. (E) Embryo carrying a mutation that eliminates four Brk binding sites (see Fig. 3B). Proper refinement is not observed in embryos carrying any of the mutant constructs.
We introduced point mutations in Brk binding sites in a zen-lacZ transgene to test if a direct interaction between Brk and the zen promoter is required for zen repression. Control gel-shift experiments showed that oligonucleotides containing the mutagenized sites did not bind recombinant Brk protein (data not shown).
Mutagenesis of B1 or B2 or both had no effect on lacZ expression (data not shown). Neither did eliminating B5 and B6 in the context of deletion Δ292–43 (Fig. 4B), nor B3 in the context of Δ476–350 (Fig. 4C). However, mutagenesis of four Brk sites (quadruple mutation in Fig. 3B) resulted in ectopic expression in the ventral ectoderm during late cellularization (Fig. 4E) similar to the pattern of zen expression in brk mutant embryos (Jazwinska et al. 1999b). These results indicate that Brk binding is essential for its repressor function. Moreover, Brk-mediated repression in the ventral ectoderm seems to depend on the cumulative contribution of Brk binding sites and not on the specific effect of any one of them.
The quadruple mutant produced an additional expression phenotype not observed in brk mutant embryos. In brk null embryos, zen expression expands into the ventral ectoderm but later refines normally (Jazwinska et al. 1999b). The wild-type zen-lacZ transgene (Fig. 4A) behaves similarly in brk mutant embryos (data not shown). In contrast, the expanded expression domain of lacZ driven by the quadruple mutant promoter did not refine, but remained broad through gastrulation (Fig. 4E). This lack of refinement is presumably not caused by the lack of Brk binding because in brk mutants, zen refines normally. Instead, it resembles the broad “nonrefining” zen-lacZ pattern seen in the two deletion mutants and the Mad site double mutant (see Fig. 4B–D). Three Brk sites mutated in the quadruple mutant also bind Mad and Medea (B1M1, B3M5, B5M7), and gel-shift experiments showed that the oligonucleotides with these mutated Brk sites do not bind Mad and Medea (data not shown). This indicates that the lack of Mad/Medea binding to the sites shared with Brk resulted in loss of refinement in the zen-lacZ quadruple mutant.
Brk is a potent repressor of zen and dpp
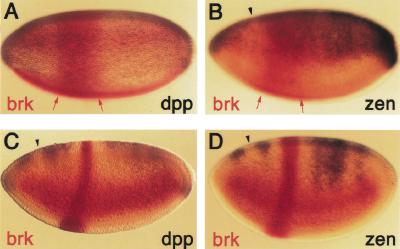
The activation of zen by Mad/Medea and its repression by Brk takes place in adjacent tissues of the embryo, the dorsal and ventral ectoderm, respectively. To test if ectopic expression of brk in the dorsal ectoderm of the embryo is sufficient to repress zen transcription, we used the FLP/FRT system in an experimental approach designed by Kosman and Small (1997). brk misexpression was driven by eve–stripe 2, a well-characterized enhancer of the even-skipped segmentation gene. The stripe is initially wide (Fig. 5A,B, red arrows) and then refines to a single anteroposterior stripe spanning parasegment 3 (Fig. 5C,D).
Figure 5.
Brk represses dpp and zen directly. Transgenic embryos carrying the FLP-out construct eve–stripe 2-brk were hybridized with either brk and dpp (A,C) or brk and zen (B,D) antisense RNA probes. dpp and zen are repressed in the region of eve–stripe 2 (parasegment 3) as Brk protein accumulates during mid- to late cellularization (C,D), but not earlier in precellular stages (A,B). Red arrows delimit the early broad stripe 2 expression. Arrowheads point to the site where normal loss of dpp and zen expression during cellularization occurs.
brk misexpression in stripe 2 represses the expression of zen, as well as dpp, in parasegment 3 beginning about mid-cellularization (Fig. 5C,D), but not earlier (Fig. 5A,B and data not shown). The repressed region appears as wide as the initial broad domain of eve–stripe 2. This result indicates that ectopic Brk represses the zen promoter directly. An indirect mechanism by which Brk represses dpp, which would then result in loss of zen maintenance, is unlikely because the effects of ectopic Brk on dpp and zen happen simultaneously (Fig. 5A,B).
Brk and Smads compete for DNA binding
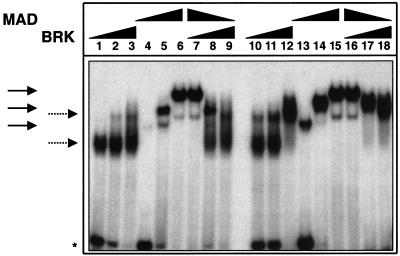
The observations that Smads can bind Brk sites and that ectopic Brk can repress zen in the presence of Smads indicated a direct competition mechanism between Smads and Brk. This could occur during mid-cellularization when Dpp maintains zen in the dorsal region while Brk represses zen in ventral regions (Jazwinska et al. 1999b). We tested the feasibility of such a mechanism in vitro by gel-shift competition assays. Oligonucleotides spanning Brk/Mad/Medea binding sites were incubated with different amounts of recombinant Brk and Mad proteins (Fig. 6). Two different sites were tested, B4M6 (lanes 1–9) and B5M7 (lanes 10–18). Increasing amounts of each of the proteins alone formed increasing amounts of complexes with different nonoverlapping mobilities (Mad complexes, solid arrows; Brk complexes, dotted arrows). When both proteins were present in the reaction, the outcome depended on their relative amounts and affinities to the corresponding oligonucleotide. At high Mad and low Brk concentrations, the complexes had mobilities of Mad alone (lanes 7 and 16). At high Brk and low Mad concentrations, the outcome was reversed and the complexes had mobilities of Brk alone (lanes 9 and 18). At intermediate concentrations, complexes with mobilities of both proteins were present (lanes 8 and 17). The formation of new complexes with different mobilities was not observed, indicating that simultaneous binding of the two proteins to the same DNA does not occur.
Figure 6.
Mad and Brk compete in vitro for binding to DNA. Electrophoretic mobility shift assay (EMSA) of complexes formed by 2 ng, 10 ng, and 50 ng (lanes 1,2,3 and 10,11,12) of Brk; 20 ng, 100 ng, and 500 ng (lanes 4,5,6 and 13,14,15) of Mad; and 2 ng + 500 ng, 10 ng + 100 ng, and 100 ng + 20 ng of Brk and Mad, respectively (lanes 7,8,9 and 16,17,18) with 0.2 ng of labeled oligonucleotides spanning B5/M7 (lanes 1–9) and B4/M6 (lanes 10–18) sites. Solid and dotted arrows indicate the positions of the Mad and Brk complexes, respectively. The asterisk indicates the position of free DNA.
Incubation of the oligonucleotide for which Mad had a higher affinity (cf. lanes 4 and 13) with intermediate concentrations of both proteins resulted in the formation of more Mad complexes, and vice versa (Fig. 6, cf. lanes 8 and 17). Other oligonucleotides spanning Brk/Mad/Medea binding sites were tested with the same results (data not shown). It can be concluded that, under the conditions used, Mad and Brk do not bind simultaneously to DNA; rather, they compete and replace each other depending on their relative concentrations and affinities for a particular binding site.
Discussion
Regulation of zen by Dpp morphogenic activity
zen has a dynamic expression pattern in the early embryo that is regulated by many factors. The initial, broad dorsal-on/ventral-off pattern in the syncytial blastoderm results from activation by a ubiquitous activator and repression in the ventral half of the embryo by the Dl morphogen (Rushlow et al. 1987; Doyle et al. 1989). The experiments described here provide evidence that zen expression during cellularization is also regulated by an interplay of negative and positive factors. zen activation becomes dependent on Dpp signaling in the dorsal ectoderm, whereas Brk protein represses zen in the ventral ectoderm. Our results further show that Dpp also is required for the refinement of zen into a narrow domain. Furthermore, the two processes involve different levels of signaling. Maintenance requires at least some intermediate level of Dpp activity whereby the more Dpp activity, the more zen transcription (Fig. 1E,G). Refinement, however, requires the highest level of signaling, because strong zen expression in the dorsal-most cells is not observed even in the weakest dpp mutant (Fig. 1E,F).
Examination of p-Mad staining in wild-type embryos also indicates that maintenance and refinement require different levels of signaling. Only the highest p-Mad levels in the dorsal-most five to six nuclei are capable of driving zen transcription during late cellularization. The lower levels present in the three to four lateral nuclei to either side are not sufficient to activate zen (Fig. 2C,D), although earlier they were sufficient for its maintenance (Fig. 2A,B). This indicates that maintenance may involve the contribution of an additional activator, perhaps the same ubiquitous activator that initiates zen earlier.
Later during refinement, p-Mad at peak levels is sufficient to up-regulate zen. Interestingly, the Dpp target gene ush is expressed in a broader domain than refined zen that includes the three to four lateral nuclei (Jazwinska et al. 1999b). This indicates that ush can be activated by a lower level of signaling than refined zen and that the high-level class of Dpp target genes can be further subdivided.
It has been shown before that the threshold target gene response to Drosophila morphogens Bcd and Dl is determined by their concentrations in the nucleus (Driever et al. 1989; Roth et al. 1989; Struhl et al. 1989). The difference between Bcd and Dl and Dpp is that because the former are transcription factors, their concentrations can be measured directly by nuclear target promoters whereas Dpp signals must be transduced and possibly modulated before reaching the nucleus. The difference, however, might not be consequential. It was shown recently in Xenopus that increasing the activated Activin receptor threefold results in a proportional increase of nuclear Smads (Shimizu and Gurdon 1999), providing evidence that the concentration of nuclear signal transducers is a readout of the extracellular ligand concentration or activity.
We can make similar conclusions from our genetic experiments. Because the amount of p-Mad depends on the amount of Dpp activity, the simplest explanation is that the zen promoter responds to differences in Dpp activity by measuring the level of nuclear Smads. Such a conclusion is consistent with the presence of multiple Mad/Medea binding sites and our mutagenesis analysis. Deletion of only two Mad/Medea sites resulted in the loss of refined expression (Fig. 4D); therefore, most if not all of the Smad binding sites are required for this function, as are the peak levels of p-Mad activity (because weak dpp mutants do not refine). However, maintenance was not affected, possibly because several sites remain intact and this function does not require full p-Mad activity. That the zen promoter measures the level of nuclear Smads also explains the broad dorsolateral pattern of both p-Mad immunostaining and zen expression in sog− embryos. In the absence of inhibition by Sog, Dpp continues to signal, and p-Mad can accumulate in the dorsolateral region of the embryo and induce zen expression (Fig. 2L).
zen regulation by Brk and Smads
The experiments presented here show that Mad/Medea and Brk regulate zen by binding to separated and overlapping DNA binding sites. There are 10 Mad/Medea and 6 Brk binding sites in the zen promoter, 5 of which are shared, indicating duality in their function. Indeed, the results from mutagenesis of the zen promoter show that the shared sites mediate both Brk and Mad/Medea functions.
Five Brk and nine Smad binding sites are clustered in the zen proximal regulatory element over about 600 bp with spacing not exceeding 120 bp. This organization is similar to that of several well-studied enhancers from Drosophila (Small et al. 1991; Hoch et al. 1992; Ip et al. 1992). These enhancers are activated by a variety of transcriptional activators and repressed by short-range repressors such as Snail (Sna; Gray et al. 1994), Knirps (Kni; Arnosti et al. 1996), and Krüppel (Kr; Gray and Levine 1996). All three of these repressors are DNA-binding proteins that can inhibit activator function when they are bound not further than 150 bp away from the activator binding site. It was shown that they all contain a short stretch of amino acids, P-DLS-K, that is required for recruitment of the corepressor dCtBP (Nibu et al. 1998). Our analysis of zen regulation indicates that Brk also may be a short-range repressor. It is a DNA-binding protein and contains a PMDLSG domain (Jazwinska et al. 1999a). Preliminary in vitro experiments showed that Brk interacts with dCtBP (N. Kirov, unpubl.); however, embryos devoid of dCtBP activity do not ectopically express zen and dpp (C. Rushlow, unpubl.), indicating that dCtBP is dispensable for Brk repression and other corepressors interact with Brk, or that Brk repression of these targets does not require additional factors.
The identity of the ubiquitous transcriptional activator that activates zen in the dorsal ectoderm during precellular stages and early cellularization remains elusive. It is possible that this activator interacts with Smads to enhance transcription of zen at a time when p-Mad levels are low. Also, Brk represses the ubiquitous activator, because zen becomes ectopically expressed in brk mutants (Jazwinska et al. 1999b). Thus far, our deletion analysis of the zen promoter has not uncovered any sequences that might interact with this putative activator. It is possible that these sequences are redundant and scattered over the entire promoter and may in fact overlap with Smad and/or Brk binding sites.
A competition model for zen transcriptional regulation
In the cellularizing embryo, Dpp and Brk activities overlap in the lateral-most region. Here Dpp and Brk function to set thresholds of response for target genes such as zen and pnr (Jazwinska et al. 1999b). In this same region, Dpp signaling negatively regulates brk expression. Similarly, in the wing disc, the Brk expression domain overlaps with that of the Dpp target gene omb (Jazwinska et al. 1999a) in the region where activated p-Mad is present (Tanimoto et al. 2000). It was proposed that a dual mechanism whereby Dpp can simultaneously down-regulate Brk repressor levels and antagonize its function on target gene promoters would be very efficient in establishing sharp threshold responses (Jazwinska et al. 1999b). Based on our experiments, we suggest a molecular model to explain mechanistically the antagonizing activities of Brk and Smads. We propose that they are involved in direct competition for binding to shared binding sites on target promoters. Thus, it is the balance of their opposing activities that determines the transcriptional state of the target genes. Two sets of experiments support this model. First, ectopic expression of Brk in eve–stripe 2 abolished zen expression in those cells (Fig. 5C).
The elevated level of Brk in the stripe was therefore sufficient to repress the zen promoter even in the presence of activated Smads. The possibility that zen was repressed indirectly through Brk-mediated repression of dpp is highly unlikely because there was no delay in zen repression. Second, our in vitro competition experiments also support the model. Especially revealing is the fact that the outcome of competition depends on the relative concentrations of both proteins and their binding affinities (Fig. 6, lanes 8 and 17). Competitive mechanisms have been proposed to operate on many promoters where mutually exclusive DNA-binding factors are involved, and, in some instances, DNA-binding assays similar to ours were used to show competition for binding between activator and repressor proteins (Small et al. 1991; Hoch et al. 1992; Rahuel et al. 1992; Genetta et al. 1994). For example, bHLH proteins compete with a zinc-finger repressor for E-box binding in the immunoglobulin heavy chain enhancer (Genetta et al. 1994).
Implications for the regulation of Dpp target genes
The findings presented here provide a framework for further study of the mechanisms of regulation of Dpp morphogen targets. zen is the only one of the known Dpp target genes that responds to two threshold activities: low (during early to mid-cellularization) and high (during late cellularization). Based on our results and the proposed competition mechanism for activation and repression of the zen promoter, we can make predictions about the organization of the regulatory elements of the other Dpp target genes. High-level targets such as ush strongly depend on high levels of Smads, and their regulatory elements may have many, and possibly closely packed, Smad binding sites.
Low-level targets such as omb (Grimm and Pflugfelder 1996) in the wing imaginal disc may be repressed by Brk binding to their regulatory sequences (Jazwinska et al. 1999a,b). The spatial domains of expression of the intermediate targets such as pnr in the embryo and sal (de Celis et al. 1996) in the wing disc, which are dependent on both Dpp signaling and Brk repression (Jazwinska et al. 1999a,b), might be determined by the net balance of positive and negative inputs. Interestingly, this type of mechanism can result in expression domains that vary largely in size and may result in even broader domains than the low-level targets. An example is the vg gene. In third-instar imaginal wing discs, vg is expressed in a broader domain than omb (Kim et al. 1996, 1997). Its expression along the anterior-posterior boundary in the wing pouch is activated by the quadrant enhancer that contains Mad binding sites essential for activation (Kim et al. 1997). At the same time, vg is repressed by Brk (Campbell and Tomlinson 1999). However, the essential Smad binding sites do not match the Brk binding sites (N. Kirov, unpubl.), like many of the Smad sites in the zen promoter, suggesting that they will have no or low affinity for the Brk protein. Neither are there strong zen-like Brk binding sites in the quadrant enhancer (sequence from Kim et al. 1997). Its broad expression domain could then be explained if the positive inputs from Smads, enhanced by signals from the dorsoventral boundary (Kim et al. 1996), are able to overcome Brk repression far from the Dpp source.
Further studies of the arrangement, affinities, and numbers of repressor and activator sites in Dpp target promoters will determine to what extent the different thresholds of responses to the Dpp morphogen activity are shaped by a simple balance of positive and negative transcriptional inputs.
Materials and methods
Drosophila stocks
dpp hypomorphic alleles (Wharton et al. 1993): dpphr4, dpphr27, dppH94; dpp amorphic allele: dppH46. dpphr4 is balanced over SM6, eve-lacZ, whereas the remaining alleles are balanced over CyO23. sog allele (Biehs et al. 1996): sogYS06 /FM7c, ftz-lacZ.
In situ hybridization and antibody staining
Wild-type and mutant embryos were fixed, hybridized with zen or lacZ RNA antisense probes, stained (reagents obtained from Roche Molecular Biochemicals), dehydrated, prepared for sectioning, and mounted (araldite obtained from Polysciences) as described in Roth et al. (1989). Anti-pMad polyclonal antibodies were kindly provided by P. ten Dijke and used at a final dilution of 1:1000 in PBS. lacZ expression was detected using rabbit anti-βGal (Cappel) antibodies. Secondary antirabbit antibody staining was performed using the Vectastain ABC kit. Embryo sections were photographed using DIC optics on a Nikon FX-A microscope.
eve–stripe 2 misexpression
The full-length brk cDNA (Jazwinska et al. 1999a) was cloned into the eve–stripe 2 misexpression vector (Kosman and Small 1997). The construct contains two copies of the 480-bp eve–stripe 2 enhancer, the 3′ transcription termination signal from the hsp70 gene flanked by two FLP recombination targets (FRTs), and the brk cDNA. Upon recombination, brk expression is driven by the eve–stripe 2 enhancer. Transgenic flies carrying this construct were kindly provided by S. Small. Two independent lines were each crossed to a transgenic line homozygous for a P(ry+), β2-tubulin-FLP insertion to obtain males that carried both constructs.
Embryos were then collected from crosses between these males and yw females and stained by in situ hybridization using antisense RNA probes. The double-label experiments were performed as described previously (Kosman and Small 1997) using a combination of brk labeled with fluorescein-UTP and either zen or dpp labeled with digoxygenin-UTP. Embryos were mounted in 50% glycerol and photographed as above.
Expression of Brk, Mad, and Medea
The DNA-binding domain of Brk was mapped in the N-terminal 100 amino acids by truncation and point mutation analysis (data not shown). GST–Brk fusion protein used in our experiments contained the N-terminal 266 amino acids. It was cloned by introduction of an NdeI site in the first codon of Brk, excision of a 793-bp NdeI–BamHI fragment from the mutagenized brk cDNA, and blunt-end ligation into the filled-in BamHI site of the pGEX-4T-2 vector (Pharmacia). Control experiments showed that its DNA-binding activity is indistinguishable from that of the full-length protein (data not shown). Expression plasmids encoding Mad–GST (MadN) and Medea–GST fusion proteins containing the N-terminal MH1 domains were obtained from A. Laughon (Kim et al. 1997) and M. Frasch (Xu et al. 1998), respectively. The expression and purification of the recombinant proteins were performed as described previously (Kirov et al. 1993). The concentration of the isolated proteins was determined by SDS-PAGE after staining with Coomassie R-250, together with defined amounts of bovine serum albumin.
In vitro DNA-binding assays
The electrophoretic mobility-shift assays were performed as described previously (Kirov et al. 1993) except that the electrophoresis was run at room temperature. The sequences of the oligonucleotides spanning B5/M7 and B4/M6 used in the competition experiments were: 5′-TCTCTCACGCAACGCC AGATCCTGGCAGGA and 5′-CTGGCGAGACTCCCGGCT CTTGCGGCCCAG. The labeled oligonucleotides were added to the reactions containing corresponding amounts of Brk, Mad, or both proteins and incubated for 30 min at room temperature before loading on the gel. DNase I footprint analyses were performed as described previously (Kirov et al. 1993).
In vitro mutagenesis and transgenic analysis
Mutations were created in a plasmid containing the zen 1.6-kb full-length promoter, untranslated leader, and ATG fused in frame to lacZ (Doyle et al. 1989). Deletions were designed based on convenient restriction sites or highly homologous sequences between Drosophila melanogaster and D. virilis (N. Kirov, unpubl.). The Δ292–43 deletion was made by removing an AccI restriction fragment and religating the plasmid. The remaining deletions and point mutations were made using the Mutagene in vitro mutagenesis kit (BioRad).
zen–lacZ constructs were subcloned into the Casper transformation vector as described previously (Doyle et al. 1989). At least three transformant lines for each construct were tested.
Acknowledgments
We thank Kristi Wharton for dpp fly stocks and Peter ten Dijke for the anti-Smad 1 antibodies. We are indebted to Serena Silver, Colleen Noviello, and Lilly Sehgal for help in analyzing transformant embryos, and Jodi Meltzer and Steve Small for the eve–stripe 2–Brk transformants. We thank Siegfried Roth, Claude Desplan, and Steve Small for critical reading of the manuscript and many helpful discussions, and Laurel Raftery for sharing unpublished results. This work was supported by a grant from the National Science Foundation, NSF-IBN-9816881.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL car2@nyu.edu; FAX (212) 995-4015.
E-MAIL nk2@nyu.edu; FAX (212) 995-4015.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.861401.
References
- Arnosti DN, Gray S, Barolo S, Zhou J, Levine M. The gap protein Knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J. 1996;15:3659–3666. [PMC free article] [PubMed] [Google Scholar]
- Ashe HL, Levine M. Local inhibition and long-range enhancement of Dpp signal transduction by Sog. Nature. 1999;398:427–431. doi: 10.1038/18892. [DOI] [PubMed] [Google Scholar]
- Ashe HL, Mannervik M, Levine M. Dpp signaling thresholds in the dorsal ectoderm of the Drosophila embryo. Development. 2000;127:3305–3312. doi: 10.1242/dev.127.15.3305. [DOI] [PubMed] [Google Scholar]
- Biehs B, François V, Bier E. The Drosophila short gastrulation gene prevents Dpp from autoactivating and suppressing neurogenesis in the neuroectoderm. Genes & Dev. 1996;10:2922–2934. doi: 10.1101/gad.10.22.2922. [DOI] [PubMed] [Google Scholar]
- Campbell G, Tomlinson A. Transducing the Dpp morphogen gradient in the wing of Drosophila: Regulation of Dpp target genes by brinker. Cell. 1999;96:553–562. doi: 10.1016/s0092-8674(00)80659-5. [DOI] [PubMed] [Google Scholar]
- Cubbada Y, Heitzler P, Ray RP, Bourouis M, Ramain P, Gelbart W, Simpson P, Haenlin M. u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes & Dev. 1997;11:3083–3095. doi: 10.1101/gad.11.22.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cellis JF, Bario R, Kafatos FC. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature. 1996;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- Doyle HJ, Harding K, Hoey T, Levine M. Transcripts encoded by a homeobox gene are restricted to dorsal tissues of Drosophila embryos. Nature. 1986;323:76–79. doi: 10.1038/323076a0. [DOI] [PubMed] [Google Scholar]
- Doyle HJ, Kraut R, Levine M. Spatial regulation of zerknüllt: A dorsal-ventral patterning gene in Drosophila. Genes & Dev. 1989;3:1518–1533. doi: 10.1101/gad.3.10.1518. [DOI] [PubMed] [Google Scholar]
- Driever W, Thoma G, Nusslein-Volhard C. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity sites for the bicoid morphogen. Nature. 1989;340:363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- Eresh S, Riese J, Jackson DB, Bohman D, Bienz M. A CREB-binding site as a target for decapentaplegic signaling during Drosophila endoderm induction. EMBO J. 1997;16:3912–3922. doi: 10.1093/emboj/16.8.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Anderson KV. decapentaplegic acts as a morphogen to organize dorsal-ventral pattern in the Drosophila embryo. Cell. 1992;71:451–461. doi: 10.1016/0092-8674(92)90514-d. [DOI] [PubMed] [Google Scholar]
- Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box binding repressor by basic helix-loop-helix proteins: Implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Levine M. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes & Dev. 1996;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes & Dev. 1994;8:1829–1838. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- Grimm S, Pflugfelder GO. Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science. 1996;271:1601–1604. doi: 10.1126/science.271.5255.1601. [DOI] [PubMed] [Google Scholar]
- Hoch M, Gerwin N, Taubert H, Jäckle H. Competition for overlapping sites in the regulatory region of the Drosophila gene Krüppel. Science. 1992;256:94–97. doi: 10.1126/science.1348871. [DOI] [PubMed] [Google Scholar]
- Holley SA, Jackson PD, Sasai Y, Lu B, De Robertis EM, Hoffmann FM, Ferguson EL. A conserved system for dorsal-ventral patterning in insects and vertebrates involving sog and chordin. Nature. 1995;376:249–253. doi: 10.1038/376249a0. [DOI] [PubMed] [Google Scholar]
- Ip TY, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes & Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Kirov N, Wieschaus E, Roth S, Rushlow C. The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell. 1999a;96:563–573. doi: 10.1016/s0092-8674(00)80660-1. [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Rushlow C, Roth S. The role of brinker in mediating the graded response to Dpp in early Drosophila embryos. Development. 1999b;126:3323–3334. doi: 10.1242/dev.126.15.3323. [DOI] [PubMed] [Google Scholar]
- Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- Jiang J, Cai H, Zhou Q, Levine M. Conversion of a dorsal-dependent silencer into an enhancer: Evidence for dorsal corepressors. EMBO J. 1993;12:3201–3209. doi: 10.1002/j.1460-2075.1993.tb05989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen H, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Kirov N, Zhelnin L, Shah J, Rushlow C. Conversion of a silencer into an enhancer: Evidence for a co-repressor in dorsal-mediated repression in Drosophila. EMBO J. 1993;12:3193–3199. doi: 10.1002/j.1460-2075.1993.tb05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Small S. Concentration-dependent patterning by an ectopic expression domain of the Drosophila gap gene knirps. Development. 1997;124:1343–1354. doi: 10.1242/dev.124.7.1343. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Struhl G. Morphogens, compartments, and pattern: Lessons from Drosophila. Cell. 1996;85:951–961. doi: 10.1016/s0092-8674(00)81297-0. [DOI] [PubMed] [Google Scholar]
- Marqués G, Musacchio M, Shimell MJ, Wunnenberg-Stapleton K, Cho KWY, O'Connor MB. Production of Dpp activity gradient in the early Drosophila embryo through the opposing actions of the Sog and Tld proteins. Cell. 1997;91:417–426. doi: 10.1016/s0092-8674(00)80425-0. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Massagué J, Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Blain SW, Lo RS. TGF-β signaling in growth control, cancer, and heritable disorders. Cell. 2000;102:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Minami M, Kinoshita N, Kamoshida Y, Tanimoto H, Tabata T. brinker is a target of Dpp in Drosophila that negatively regulates Dpp-dependent genes. Nature. 1999;398:242–246. doi: 10.1038/18451. [DOI] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. dCtBP mediates transcriptional repression by Knirps, Krüppel and Snail in the Drosophila embryo. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson U, Izumi H, Souchelnytskyi S, Itoh S, Grimsby S, Engstrom U, Heldin CH, Funa K, ten Dijke P. The L45 loop in type I receptors for TGF-β family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 1998;434:83–87. doi: 10.1016/s0014-5793(98)00954-5. [DOI] [PubMed] [Google Scholar]
- Podos SD, Ferguson EL. Morphogen gradients: New insights from DPP. Trends Genet. 1999;15:396–402. doi: 10.1016/s0168-9525(99)01854-5. [DOI] [PubMed] [Google Scholar]
- Raftery L, Sutherland D. TGF-β family signal transduction in Drosophila development: From Mad to Smads. Dev Biol. 1999;210:251–268. doi: 10.1006/dbio.1999.9282. [DOI] [PubMed] [Google Scholar]
- Rahuel C, Vinit MA, Lemarchandel V, Cartron JP, Romeo PH. Erythroid-specific activity of the glycophorin B promoter requires GATA-1 mediated displacement of a repressor. EMBO J. 1992;11:4095–4102. doi: 10.1002/j.1460-2075.1992.tb05502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RP, Arora K, Nusslein-Volhard C, Gelbart WM. The control of cell fate along the dorsal-ventral axis of the Drosophila embryo. Development. 1991;113:35–54. doi: 10.1242/dev.113.1.35. [DOI] [PubMed] [Google Scholar]
- Roth S, Stein D, Nusslein-Volhard C. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell. 1989;59:1189–1202. doi: 10.1016/0092-8674(89)90774-5. [DOI] [PubMed] [Google Scholar]
- Rushlow C, Levine M. Role of the zerknüllt gene in dorsal-ventral pattern formation in Drosophila. Adv Genet. 1990;27:277–307. doi: 10.1016/s0065-2660(08)60028-0. [DOI] [PubMed] [Google Scholar]
- Rushlow CA, Frasch M, Doyle H, Levine M. Maternal regulation of zerknüllt: A homeobox gene controlling differentiation of dorsal tissues in Drosophila. Nature. 1987;330:583–586. doi: 10.1038/330583a0. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang Y-F, Jayaraman L, Yang H, Massagué J, Pavletich N. Crystal structure of a Smad MH1 domain bound to DNA: Insights on DNA-binding in TGF-β signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Gurdon JB. A quantitative analysis of signal transduction from activin receptor to nucleus and its relevance to morphogen gradient interpretation. Proc Natl Acad Sci. 1999;96:6791–6796. doi: 10.1073/pnas.96.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Brose M, Treisman J, Desplan C. Synergy between the Hunchback and Bicoid morphogens is required for anterior patterning in Drosophila. Cell. 1994;78:855–865. doi: 10.1016/s0092-8674(94)90622-x. [DOI] [PubMed] [Google Scholar]
- Small S, Kraut R, Hoey R, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes & Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- St. Johnson RD, Gelbart WM. decapentaplegic transcripts are localized along the dorsal-ventral axis of the Drosophila embryo. EMBO J. 1987;6:2785–2791. doi: 10.1002/j.1460-2075.1987.tb02574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Struhl K, MacDonald PM. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- Sutherland DJ, Raftery LA. 41st Annual Drosophila Research Conference. 533A. Pittsburgh, PA. 2000. Direct evidence for a DPP activity gradient in embryonic D-V patterning. [Google Scholar]
- Tanimoto H, Itoh S, ten Dijke P, Tabata T. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol Cell. 2000;5:59–71. doi: 10.1016/s1097-2765(00)80403-7. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Miyazono K, Heldin C. Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem Sci. 2000;25:64–70. doi: 10.1016/s0968-0004(99)01519-4. [DOI] [PubMed] [Google Scholar]
- Wharton KA, Ray RP, Gelbart WM. An activity gradient of decapentaplegic is necessary for the specification of dorsal pattern elements in the Drosophila embryo. Development. 1993;117:807–822. doi: 10.1242/dev.117.2.807. [DOI] [PubMed] [Google Scholar]
- Wilson PA, Lagna G, Suzuki A, Hemmati-Brivanlou A. Concentration-dependent patterning of the Xenopus ectoderm by BMP4 and its signal transducer Smad1. Development. 1997;124:3177–3184. doi: 10.1242/dev.124.16.3177. [DOI] [PubMed] [Google Scholar]
- Winick J, Abel T, Leonard MW, Michelson AM, Chardon-Loriaux I, Holmgren RA, Maniatis T, Engel JD. A GATA family transcription factor is expressed along the embryonic dorsoventral axis in Drosophila melanogaster. Development. 1993;119:1055–1065. doi: 10.1242/dev.119.4.1055. [DOI] [PubMed] [Google Scholar]
- Xu S, Yin Z, Hudson JB, Ferguson EL, Frasch M. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes & Dev. 1998;12:2354–70. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]