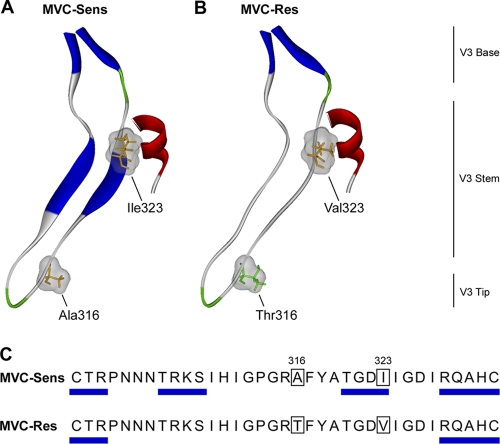
Fig. 5.
Structural analysis of MVC resistance mutations in the V3 loop. (A and B) Ribbon representations of the V3 loop regions of the MVC-Sens (A) and MVC-Res (B) gp120 proteins docked to the CCR5 N-terminal sulfopeptide. Secondary structure was predicted using the method of Kabsch and Sander (33) and was color coded as follows: blue, beta sheets; red, helices; green, coils; gray, disordered regions. Residues at positions 316 and 323 (HIVHXB2 numbering), shown by mutagenesis studies to confer MVC resistance on MVC-Res Env (76), are colored according to polarity (orange, nonpolar; green, polar), with their molecular surfaces shown in gray. (C) The predicted beta sheet structures are annotated by blue rectangles on the amino acid sequence alignment of the MVC-Sens and MVC-Res V3 loops.