Abstract
The nonstructural protein 3 (NS3) helicase/protease is an important component of the hepatitis C virus (HCV) replication complex. We hypothesized that a specific β-strand tethers the C terminus of the helicase domain to the protease domain, thereby maintaining HCV NS3 in a compact conformation that differs from the extended conformations observed for other Flaviviridae NS3 enzymes. To test this hypothesis, we removed the β-strand and explored the structural and functional attributes of the truncated NS3 protein (NS3ΔC7). Limited proteolysis, hydrodynamic, and kinetic measurements indicate that NS3ΔC7 adopts an extended conformation that contrasts with the compact form of the wild-type (WT) protein. The extended conformation of NS3ΔC7 allows the protein to quickly form functional complexes with RNA unwinding substrates. We also show that the unwinding activity of NS3ΔC7 is independent of the substrate 3′-overhang length, implying that a monomeric form of the protein promotes efficient unwinding. Our findings indicate that an open, extended conformation of NS3 is required for helicase activity and represents the biologically relevant conformation of the protein during viral replication.
INTRODUCTION
Nonstructural protein 3 (NS3) is an essential member of the hepatitis C virus (HCV) replication complex (2, 33). It is a bifunctional enzyme that contains a serine protease domain within the N-terminal third of the protein and a nucleic acid-stimulated NTPase/helicase domain within the C-terminal two-thirds of the protein (40). Both enzymatic activities are critical for HCV replication. In the presence of the viral NS4A cofactor protein, the NS3 protease activity cleaves and releases downstream viral proteins from the precursor polyprotein (13). Furthermore, the NS3 protease represses the host innate immune response by cleaving cellular proteins such as TRIF and MAVS, thereby preventing a signaling cascade that leads to an antiviral cellular response (17, 24, 32, 44, 51). In addition to its protease activity, NS3 displays robust helicase activity (46). NS3 helicase activity is essential for replication of the viral RNA genome (21), potentially functioning together with the NS5B polymerase during viral replication (16). NS3 also participates in the intracellular assembly and packaging of infectious virus particles (30). Given its multiple roles throughout the viral life cycle, NS3 is an important target for antiviral drug discovery against HCV (5).
The NS3 helicase (NS3hel) domain belongs to the DExH/D subgroup of DNA and RNA helicases within helicase superfamily 2 (37, 52). Members of this family contain a core helicase structure consisting of two RecA-like folds (domains 1 and 2) arranged in tandem. Together with these two RecA-like domains, NS3hel has a third domain (domain 3) that forms a single-stranded DNA/RNA binding groove (Fig. 1A) (20, 29). The NS3hel construct has been studied extensively and displays modest helicase activity in isolation. However, a unique feature of NS3 that distinguishes it from its other family members is the covalent attachment of the protease domain. Previous work has demonstrated a strong functional interdependence in the enzymatic activities of the protease and helicase domains (7, 8). The protease domain not only promotes direct and functional binding of the RNA substrate, but it also improves the translocation stepping efficiency of the helicase (8, 39). Therefore, full-length NS3 is an attractive target for the development of antiviral strategies.
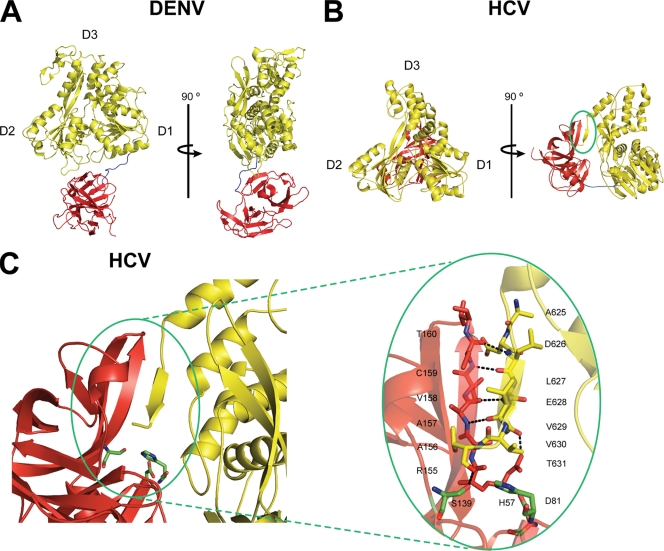
Fig. 1.
Domain orientations of DENV NS3 and HCV NS3 proteins. (A) The NS3 proteins from DENV (Protein Data Bank [PDB] no. 2VBC) and (B) HCV (PDB no. 1CU1) are shown in cartoon form, each with a 90° rotation. The protease and helicase domains are colored in red and yellow, respectively. Subdomains of the helicase domains are labeled. The linker region connecting the two domains is colored in blue. Note that the protease domain of DENV NS3 is located “below” the ATP binding pocket, while that of HCV NS3 is located “behind” the ATP binding pocket. The NS3 protein from MVEV was crystallized in an extended conformation similar to DENV NS3 (data not shown) (PDB no. 2WV9). (C) The C terminus of HCV NS3 forms a β-strand and feeds back into the protease active site to form a β-sheet (circled in green). The catalytic triad residues of the protease domain are shown as green sticks. Figures were generated by PyMol.
While previous studies have provided important insights into the mechanism of RNA unwinding by NS3 (11, 34, 43), we know very little about the structural rearrangements that occur when NS3 switches between its protease and helicase activities. One way NS3 might regulate its functional activities is through global conformational changes. An increasing body of evidence suggests that NS3 adopts additional conformations that differ from the one observed in the crystal structure of the full-length enzyme, which captured a snapshot of the protein performing a protease cleavage event in cis (53). One clue to the existence of alternative conformations came from early kinetic data showing that NS3 requires an extended incubation time with RNA substrates prior to reaching its maximal unwinding activity (36), suggesting a rate-limiting conformational change. Second, the structures of NS3 proteins from other Flaviviridae family members show that the biologically relevant conformation is an extended form where the protease domain sits beneath the helicase domain (4, 27, 28). These extended conformations contrast markedly with the compact conformation seen in the crystal structure of full-length HCV NS3 (53). Finally, when HCV NS3 was modeled onto a membrane bilayer, the helicase domain was positioned in an impossible configuration within the membrane (9). Based on this finding, Moradpour and colleagues concluded that a significant conformational change must occur in order to orient the helicase domain toward the cytoplasm and properly position it to interact with other components of the replication complex. Thus, we sought to understand how such a conformational change might occur in HCV NS3 and determine how this change would affect the function of NS3 as a helicase.
Here, we show that the C-terminal β-strand of HCV NS3 acts as a toggle that alters the structural and functional properties of the protein. The presence or absence of this β-strand changes the global architecture of NS3 and allows the protein to adopt different conformational states. Moreover, removal of this β-strand allows the truncated protein (NS3ΔC7) to adopt an extended conformation and form functional complexes with RNA unwinding substrates faster than WT NS3. In contrast to NS3ΔC7, WT NS3 cannot react efficiently under unwinding conditions that stabilize the resulting β-sheet substructure. Finally, we demonstrate that NS3ΔC7 readily unwinds RNA with short 3′-overhang strands and behaves like a processive monomer. Taken together, we propose a model in which NS3ΔC7 adopts a conformation that is distinct from the conformation observed in the crystal structure and which represents the biologically relevant form of NS3 as an active helicase.
MATERIALS AND METHODS
Materials.
DNA oligonucleotides were obtained from Invitrogen. RNA oligonucleotides were synthesized on an automated MerMade 6 synthesizer (BioAutomation) by using phosphoramidite chemistry (phosphoramidites purchased from Glen Research) and deprotected before gel purification (48).
RNA duplex substrates.
The sequences of the RNA duplexes used in this study were described previously (18, 36). RNA duplexes were labeled with [γ-32P]ATP as described previously (6), purified from semidenaturing polyacrylamide gels (15% acrylamide, 2 M urea, 0.5× Tris-borate-EDTA [TBE]), and stored at −80°C in ME buffer (10 mM MOPS [morphoplinepropanesulfonic acid], pH 6.5, 1 mM EDTA). Duplex concentrations were determined with a NanoDrop (Thermo Scientific), and extinction coefficients were calculated from the sequences.
Protein purification.
Proteins were purified as described previously with modifications (6). Proteins were expressed in Rosetta II (DE3) cells (Novagen) with 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 20 h at 16°C. Proteins were purified with Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) and heparin affinity columns (GE Healthcare) before gel filtration over a 16/60 Superdex 200 column (GE Healthcare) equilibrated in storage buffer (10 mM HEPES, 300 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol [β-ME], pH 7.0). Monodisperse fractions were concentrated, flash frozen in liquid nitrogen, and stored at −80°C. Protein concentrations were measured in 6 M guanidinium at λ = 280 nm using an extinction coefficient of ε = 63,580 M−1 cm−1 (12).
Limited proteolysis assay.
Lyophilized carboxypeptidase Y (CPY) was obtained from Worthington Biochemical (Lakewood, NJ) and resuspended in storage buffer (100 mM MOPS, 10% glycerol, 300 mM NaCl, 5 mM β-ME, pH 6.3) to a final concentration of 1 mg/ml. WT NS3 and NS3ΔC7 proteins were diluted in this buffer to final concentrations of 300 μg/ml. Proteolysis experiments were performed at 24°C with 360 μg protein and either 0, 1, 5, or 10 μg carboxypeptidase Y. Proteolysis was monitored by quenching 15-μg protein aliquots, resolving 2.5 μg on 10% SDS-PAGE gels, and staining with Coomassie brilliant blue.
Sedimentation velocity studies.
Experiments were performed at 25°C in a buffer containing 10 mM HEPES, 150 mM NaCl, 5% glycerol, 5 mM β-ME, pH 7.0, in a Beckman Optima XL-I analytical ultracentrifuge. A four-position AN 60 Ti rotor, together with Epon 12-mm double-sector centerpieces, was used at 42,000 rpm. Radial absorption scans were measured at λ = 280 nm with a radial increment of 0.003 cm. Data analyses were performed in Sedfit 8.0 (http://www.analyticalultracentrifugation.com) (41), using a 95% confidence limit. Sedimentation coefficients at the experimental temperature, buffer density, and viscosity were corrected to standard conditions (s20, w) using the program SEDNTERP (http://jphilo.mailway.com).
Functional complex formation assay.
Functional complex formation assays were performed as previously described with modifications (36). A master mix containing 25 nM protein and 1 nM RNA duplex (final concentrations) was assembled in helicase reaction buffer (42) for 10 min on ice and then divided into 9-μl reaction aliquots. Each reaction mixture was incubated at 37°C for various amounts of time, and then 3 μl of an ATP-trap mixture (16 mM ATP and 4 μM protein trap in helicase assay buffer) was added to initiate a single cycle of unwinding. The trap was a 60-nucleotide (nt), single-stranded DNA oligonucleotide (42). We used a 34-nt, single-stranded RNA oligonucleotide as the trap when we sought to increase the final unwinding amplitude (6). Reactions were quenched after 5 min by adding 3 μl of a 5× quench buffer (42) and immediately transferred to dry ice. Duplex and single-stranded RNAs were separated on semidenaturing polyacrylamide gels (described above), visualized with a Storm820 PhosphorImager, and quantified with ImageQuant software (GE Healthcare). The fraction of unwound duplex for each time point was quantitated by the following formula: Aunw = Iss/(Iss + Ids), where Iss is the intensity of the displaced strand and Ids is the intensity of the duplex substrate. At least three experiments were performed for each kinetic analysis reported here.
Data analysis.
Data were processed with Origin 8.0 (OriginLab Corporation). Kinetic parameters describing the data for each protein and its set of unwinding substrates were determined by using global fitting methods. Rate constants and number of steps were treated as shared global parameters for each set of substrates. Unwinding amplitudes were treated as unique values for each substrate.
The data for WT NS3 were modeled to scheme 1:
using equation 1:
| (1) |
where Aunw is the unwinding amplitude at incubation time t, A0 is the apparent fraction of functional complex formed on the RNA substrate, k1 is the rate constant in which WT NS3 undergoes a conformational change from a compact (NS3c) to an extended (NS3e) conformation, and k2 is the rate constant at which WT NS3 forms a functional complex with the RNA substrate.
The data for NS3ΔC7 were modeled to scheme 2:
using equation 2:
| (2) |
where Aunw is the unwinding amplitude at incubation time t, A2 is the amplitude of the fast exponential rise in activity with the rate constant k2, and A3 is the amplitude of the decay process with rate constant k3.
Under conditions of increased glycerol and NaCl concentrations, the data were modeled to the “n-step” mechanism (26) shown in scheme 3:
where S is substrate, In is the nth intermediate state, P is the product, and k is the kinetic rate constant at each step. Data were fit using the incomplete gamma function with equation 3:
| (3) |
where Aunw is the unwinding amplitude at incubation time t, A0 is the apparent fraction of functional complex formed on the RNA substrate, n is the number of kinetic steps, and k is the observed rate constant.
Linear regression was performed with equation 4:
| (4) |
where Aunw is the unwinding amplitude, y0 is the offset, m is the slope, and x is the number of binding sites.
Processivity analysis.
The maximum unwinding amplitude for each substrate duplex was determined by fitting the data to either equation 1 or 2 for WT NS3 or NS3ΔC7, respectively. Values were plotted as a function of duplex length and fit to the equation Aunw = xPN,, where Aunw is the unwinding amplitude, x is the fraction of functional complex, P is processivity, and N is the number of kinetic steps made on the duplex. For this analysis, we used a kinetic step size of 16 bp, which had been previously determined for WT NS3 (42). We assume that NS3ΔC7 has an identical step size.
RESULTS
Differing domain orientations of Flaviviridae NS3 proteins.
When the crystal structures of NS3 proteins from HCV, dengue virus (DENV), and Murray Valley encephalitis virus (MVEV) are compared, it is clear that HCV NS3 differs dramatically from the other proteins. Specifically, a β-strand in the HCV NS3 structure clamps the protease domain next to the helicase domain, thereby creating a compact conformational state that differs from the extended conformations of DENV and MVEV (Fig. 1A). This β-strand consists of the C-terminal seven amino acids of the helicase domain, which feed into the protease active site at the N terminus of the protein (circled in Fig. 1B; see Fig. S1 in the supplemental material). The resulting β-sheet interaction between the protease and helicase domains (henceforth called the β-interaction) serves as a substrate recognition platform for proteolytic cis cleavage during the maturation of NS3. However, the β-interaction must be disrupted in order for the protease to release downstream viral proteins from the viral polyprotein. We speculated that truncation of the NS3 C terminus would uncouple the protease and helicase domains, thereby allowing the protease domain to swing away from the helicase domain and permitting NS3 to adopt an extended conformation that is similar to those of the DENV and MVEV NS3 proteins. To test this hypothesis, we created an NS3 variant (NS3ΔC7) in which we removed the C-terminal residues that comprise the β-strand tether, and we examined the behavior of this mutant protein.
The C terminus of NS3ΔC7 is solvent accessible and sensitive to protease.
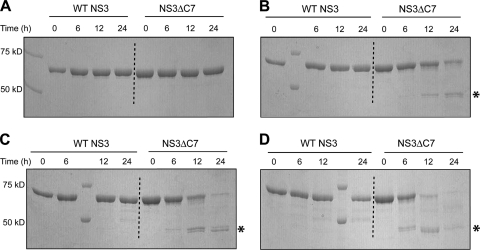
To test whether removal of the β-interaction causes the C terminus of NS3ΔC7 to become solvent accessible, we subjected both WT NS3 and NS3ΔC7 to limited proteolysis with carboxypeptidase Y (CPY) (49). In the absence of CPY, both proteins migrate as single species on an SDS-PAGE gel (Fig. 2A). In the presence of increasing amounts of CPY, WT NS3 remained insensitive to the protease (Fig. 2B to D, left), while CPY digested NS3ΔC7 with an efficiency proportional to the amount of CPY included in the reaction (Fig. 2B to D, right). These results show that WT NS3 adopts a conformation with a solvent-inaccessible C terminus, whereas the conformation of NS3ΔC7 has a solvent-exposed C terminus that renders it susceptible to CPY. The ability of CPY to digest NS3ΔC7 indicates that we have successfully eliminated the β-interaction between the protease and helicase domains.
Fig. 2.
Limited proteolysis with carboxypeptidase Y. WT NS3 (first four protein lanes) or NS3ΔC7 (last four protein lanes) was incubated with increasing amounts of CPY at 24°C. The amounts of CPY and the ratios of protein to CPY (wt/wt) are as follows: (A) 0 μg CPY; (B) 1 μg CPY, 360:1; (C) 5 μg CPY, 72:1; and (D) 10 μg CPY, 36:1. Asterisks denote stable proteolytic products. Using electrospray mass spectrometry (ES/MS), the exact masses of these proteolytic products correspond to a region within the NS3 sequence that contains a stretch of consecutive arginine and glycine residues that partially inhibit CPY activity (48) (see Fig. S1 in the supplemental material).
Hydrodynamic analysis of the global conformation of NS3ΔC7.
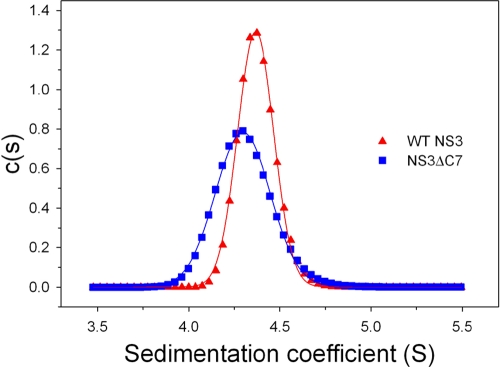
To directly examine whether WT NS3 and NS3ΔC7 have different global conformations, we performed sedimentation velocity experiments to evaluate the shapes and hydrodynamic properties of the proteins. We observe that NS3ΔC7 displays a distinct and reproducible 2.3% decrease in the sedimentation coefficient (S) relative to WT NS3, together with a broadening of the sedimentation peak (Fig. 3). For two proteins of similar molecular masses (∼1% difference), such a decrease indicates that NS3ΔC7 has a higher frictional coefficient than WT NS3 and therefore adopts a more elongated conformation. We interpret the broadening of the NS3ΔC7 sedimentation peak to reflect multiple elongated conformations as the protease domain moves freely in solution (27, 28). The presence of a single dominant peak in the data for both proteins show that the majority of the population sediments as monodisperse species in solution. These data are consistent with gel filtration results indicating that both proteins purify as stable monomers (data not shown).
Fig. 3.
Hydrodynamic analysis using sedimentation velocity. Shown is the calculated distribution c(s) versus S of WT NS3 (triangles) and NS3ΔC7 (squares). Data were acquired at a protein concentration of 5 μM, a rotor temperature of 25°C, and a rotor speed of 42,000 rpm. The sedimentation coefficients were 4.4S for WT NS3 and 4.3S for NS3ΔC7.
NS3ΔC7 forms functional complexes on RNA duplexes faster than WT NS3.
A fundamental property of helicases is the ability to bind and engage nucleic acid substrates in a productive manner that leads to strand separation. Helicases are often positively charged proteins with a high, nonspecific affinity for nucleic acids. Sequence composition, chemical identity (DNA versus RNA), or structure (single stranded versus double stranded) of the nucleic acid can influence binding specificity. Thus, direct binding assays for helicase substrate affinity may not reflect the full functional specificity of a given helicase. For example, NS3 binds nucleic acid duplexes containing either a 3′ or 5′ overhang, but it only unwinds a duplex containing a 3′ overhang (46). To monitor the bound states of NS3 that directly contribute to unwinding, we employed an activity assay that reports the rate constant at which a helicase forms functional complexes that are capable of unwinding target RNA substrates (36).
The functional complex assay begins by combining RNA duplex substrate with helicase (in excess; see Materials and Methods) and then dividing this mixture into multiple aliquots in order to vary the incubation time for each sample at the desired reaction temperature. Following the incubation period, unwinding is initiated with the simultaneous addition of ATP and a helicase “trap,” which is provided in excess to ensure single-cycle unwinding conditions. (For example, any helicase that falls off is trapped and cannot rebind.) The trap also sequesters excess helicase protein in solution from participating in unwinding after the reaction has already initiated. Each unwinding reaction is quenched after a constant reaction time, and the products are separated by PAGE (see Fig. S2 in the supplemental material). Quantification of the fraction of unwound product versus the incubation time yields the rate constant at which the bound helicase forms an activated, functional complex that unwinds RNA.
Previous studies have shown that WT NS3 requires an incubation period of 30 min with RNA duplexes at 37°C prior to achieving maximal unwinding activity (36). We reasoned that this long incubation period might reflect the time required to disrupt the β-interaction and allow conformational changes to occur within the protein-RNA complex. NS3ΔC7, lacking the β-interaction, would not require such an extended incubation step since it already adopts an extended conformation that is poised for immediate, functional binding to RNA substrates. To test this hypothesis, we measured the rate constants at which the protein constructs form functional complexes on a set of RNA unwinding substrates with progressively longer duplex lengths (36).
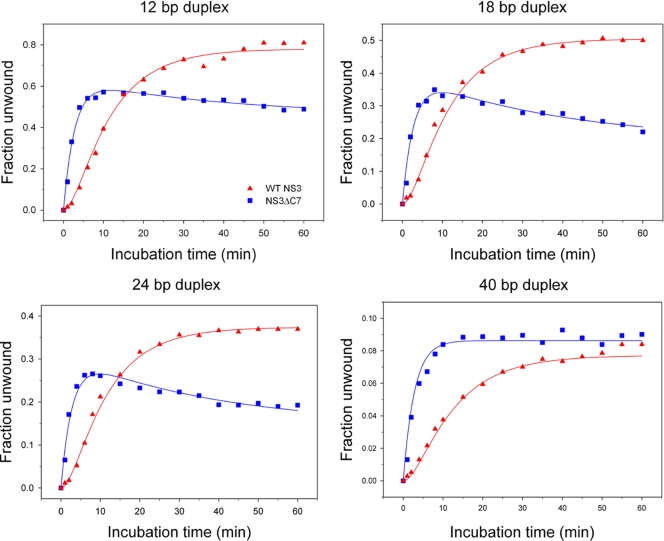
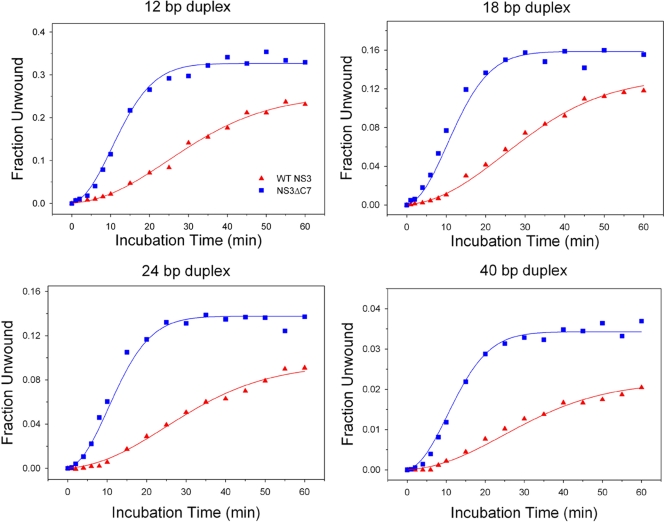
Time courses of unwound substrate versus incubation time reveal that NS3ΔC7 forms functional complexes more rapidly than WT NS3 (Fig. 4). However, the amplitude (final extent) of unwinding is generally higher for WT NS3. The data for WT NS3 display a prominent lag phase in the evolution of product for all four duplex substrates. Using global fitting methods, we fit the WT NS3 data to the mechanism shown in scheme 1 (see Materials and Methods), which includes two macroscopic rate constants: k1 is the rate constant for a putative conformational change within the NS3 protein (0.1 ± 0.01 min−1), and k2 is the rate constant at which the helicase-RNA complex adopts an activated state that proceeds immediately to unwind the duplex (0.35 ± 0.06 min−1). Such an activated state may reflect partial invasion at the duplex junction that leads to productive interactions between the helicase and RNA (22).
Fig. 4.
Functional complex formation on RNA duplexes. The fraction of RNA duplex unwound versus incubation time is plotted for WT NS3 (triangles) and NS3ΔC7 (squares). The four RNA substrates were 12, 18, 24, or 40 bp in length and contained an 18-nt, single-stranded 3′ overhang. Continuous curves represent the results of global fitting of the data using the reaction schemes described in Materials and Methods. Best fits for WT data yielded k1 = 0.1 ± 0.01 min−1 and k2 = 0.35 ± 0.06 min−1. Best fits for NS3ΔC7 data yielded k2 = 0.36 ± 0.02 min−1 and k3 = 0.03 ± 0.01 min−1.
In contrast, the data for NS3ΔC7 display a fast exponential rise in functional complex formation followed by a slow decay in activity. These data were fit using global fitting methods to scheme 2 (see Materials and Methods), which lacks the lag phase (k1) and instead includes two macroscopic rate constants: k2 is the rate constant for functional complex formation (0.36 ± 0.02 min−1), as described above, and k3 is a rate constant for the apparent decay in activity observed late in the time courses for NS3ΔC7 (0.03 ± 0.01 min−1). The decay phase likely reflects an increased instability of the NS3ΔC7 protein under the experimental conditions.
These results are consistent with a model in which WT NS3 undergoes a slow conformational change prior to forming activated complexes on RNA substrates. Once this protein isomerization has occurred (reflected by the NS3 lag phase k1), WT NS3 forms functional complexes with the same rate constant as NS3ΔC7 (k2 values are within error). Already in the activated state, NS3ΔC7 bypasses the need for a slow conformational change and proceeds quickly to form functional complexes that can unwind RNA.
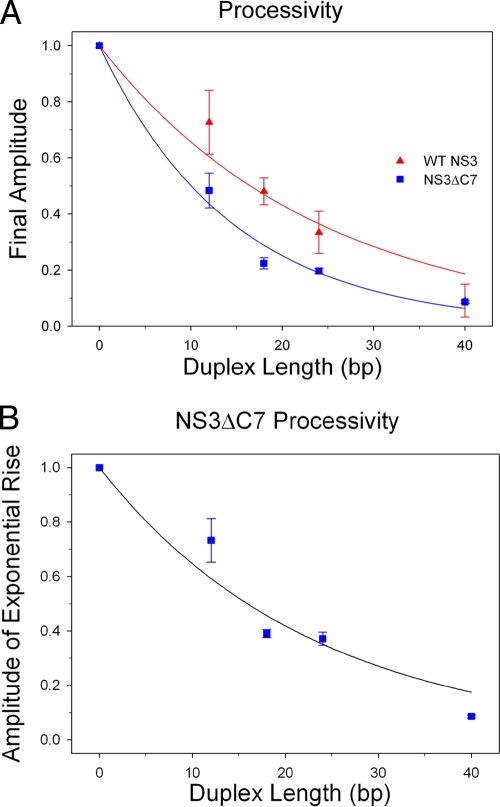
Comparative processivity of WT NS3 and NS3ΔC7.
Processivity reflects the probability that a helicase will perform a subsequent unwinding step rather than dissociate from the substrate (1, 15). Hence, a helicase of infinite processivity will unwind duplex substrates, independent of duplex length, to completion and have a processivity value equivalent to unity. A helicase of finite processivity, such as NS3, will show progressively lower unwinding amplitudes as the length of the duplex increases and have a processivity value less than unity (Fig. 4) (26). Under single-cycle conditions, these amplitudes can be used to calculate and compare the relative processivity values of the two different NS3 protein constructs. Qualitatively, it appears that NS3ΔC7 displays lower unwinding amplitudes than WT NS3 on all RNA substrates except the 40-bp duplex (Fig. 4). Based on these uncorrected final amplitudes, processivity values are P(WT NS3) = 0.50 ± 0.04 and P(NS3ΔC7) = 0.30 ± 0.02 (see Materials and Methods and Fig. 5A), indicating that NS3ΔC7 is a less processive helicase than WT NS3. However, the final unwinding amplitudes for NS3ΔC7 are the sum of two independent amplitudes that describe the exponential rise and the gradual decay in activity (scheme 2). Correction for protein decay by using only the amplitudes associated with rate constant k2, the processivity value of NS3ΔC7 increases to P(NS3ΔC7; A2) = 0.49 ± 0.05 (Fig. 5B). This finding leads us to conclude that NS3ΔC7 and WT NS3 display similar levels of helicase processivity under these unwinding conditions.
Fig. 5.
Comparative processivity measurements. WT NS3 (triangles) and NS3ΔC7 (squares) were fit by using an unwinding step size of 16 bp that had been previously determined for WT NS3. (A) The data points for NS3ΔC7 are a sum of the amplitudes that reflect the exponential rise and decay processes. Fitting of the data yields P(WT NS3) = 0.50 ± 0.04 and P(NS3ΔC7) = 0.30 ± 0.02. (B) Processivity of NS3ΔC7 using corrected amplitude values. Fitting of the data yields P(NS3ΔC7) = 0.49 ± 0.05. Error bars represent the standard deviations from a minimum of three independent experiments.
Increased glycerol concentrations inhibit functional complex formation.
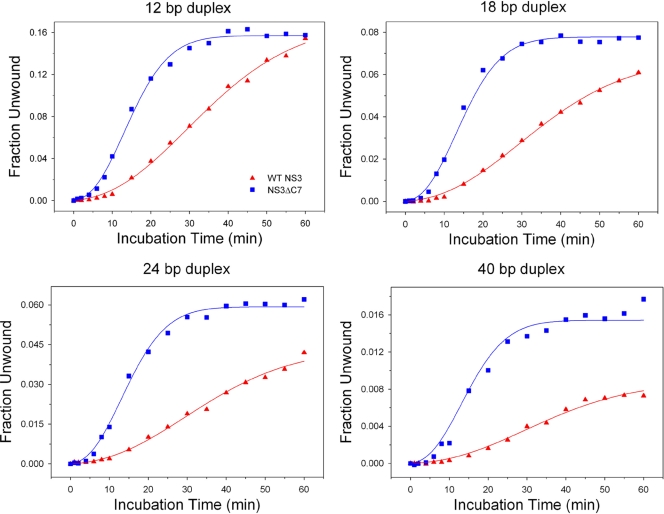
To test whether the β-interaction is inhibitory for helicase activity, we attempted to identify reaction conditions that might strengthen the interaction. We reasoned that strengthening the β-interaction would inhibit functional complex formation for WT NS3 (a significant decrease in k1), while NS3ΔC7 might be unaffected by comparison. To this end, we evaluated the effect of glycerol on the reaction. Glycerol stabilizes native protein structure by several mechanisms that are not mutually exclusive: (i) stabilization of the hydrophobic core, (ii) exclusion of glycerol from hydrophobic portions of a protein with preferential interactions with hydrophilic regions, and (iii) strengthening of hydrogen bonding networks (protein secondary structure) by preventing bulk water from acting as a hydrogen bond competitor (10, 38, 47). Increasing the glycerol concentration from 1% to 15% in our experimental conditions would be expected to perturb functional complex formation on RNA duplexes for WT NS3 by creating a larger kinetic barrier between the compact and extended conformations of the protein. WT NS3 might overcome such a barrier by requiring a longer incubation time prior to adopting the extended conformation.
In the presence of 15% glycerol, NS3ΔC7 forms functional complexes faster than WT NS3 and achieves higher unwinding amplitudes (Fig. 6). Notably, the rapid rise of activity in the NS3ΔC7 progress curves are not followed by a slow decay phase (k3) seen under standard experimental conditions, thus demonstrating the stabilizing effect of glycerol for NS3ΔC7. Glycerol caused the appearance of a small lag phase in the data for NS3ΔC7, which may reflect minor conformational changes prior to functional complex formation. Kinetic parameters were obtained by global fitting of the data to scheme 3 (see Materials and Methods; n = 2.9, k = 0.22 min−1) (26). From these results, it is clear that structural stabilization of NS3ΔC7 does not diminish its ability to adopt an activated complex and unwind RNA.
Fig. 6.
Functional complex formation in the presence of 15% glycerol. The concentration of glycerol in the reaction conditions was increased from 1% to 15%. Continuous curves represent the results of global fitting of the data for WT NS3 (triangles) and NS3ΔC7 (squares) to equation 3 as described in Materials and Methods. Best fits for WT data yielded n = 2.5 ± 0.2 and k = 0.072 ± 0.01 min−1. Best fits for NS3ΔC7 data yielded n = 2.9 ± 0.2 and k = 0.22 ± 0.003 min−1. In these experiments, the value of “n” is not implied to have a specific physical meaning and is a mathematical consequence of the hyperbolic shape of these particular curves. Note that values for n are approximately 3 in all of these experiments (including those in Fig. 7), but values for k vary greatly. Given that n remains approximately the same, we suggest that variations in k provide a direct means for comparison between the proteins. It is therefore most significant that the rate constant for NS3ΔC7 vastly exceeds that of the WT under all these conditions.
Glycerol is highly detrimental to functional complex formation by WT NS3. As expected, we observed an extended lag phase and reduced rate constant for functional complex formation (Fig. 6) (n = 2.5, k = 0.072 min−1). The reduction in rate constant suggests that glycerol stabilizes the β-interaction and prevents WT NS3 from quickly forming functional complexes. Additionally, the final unwinding amplitudes for WT NS3 are consistently lower than those for NS3ΔC7, which reflects a lower processivity for WT NS3 under these experimental conditions. Taken together, the minimal impact of glycerol on NS3ΔC7, combined with the catastrophic effect on WT NS3, supports the idea that WT NS3 is locked into a stable, inactive conformation that is sealed shut by the β-interaction.
Increased NaCl concentrations inhibit functional complex formation.
An alternative approach for stabilizing protein structure is to increase the ionic strength of the reaction buffer (3). In order to test whether the β-interaction might be stabilized under conditions of increased ionic strength, the concentration of NaCl was changed from 30 mM to 150 mM. This condition was chosen because 150 mM NaCl is the approximate salt concentration in a physiological environment. Experiments under this condition might therefore provide insights into the behavior of NS3 in vivo.
In a buffer that contains 150 mM NaCl, the activity of WT NS3 is severely inhibited at all duplex lengths (Fig. 7) (n = 2.7; k = 0.063 min−1). In contrast, NS3ΔC7 efficiently formed functional complexes and attained higher final unwinding amplitudes (Fig. 7) (n = 3.5; k = 0.22 min−1). The decay phase (k3) observed under standard experimental conditions was not evident under the high-salt conditions and demonstrates the stabilizing effect of higher NaCl concentration. Standard experimental conditions had been previously optimized to observe maximal activity by WT NS3 but not NS3ΔC7 (36). Therefore, it is significant that we have identified unwinding conditions that stabilize NS3ΔC7 and displays superior levels of functional complex formation relative to WT NS3.
Fig. 7.
Functional complex formation under high-salt conditions of 150 mM NaCl. The concentration of NaCl in the reaction conditions was increased from 30 mM to 150 mM. Continuous curves represent the results of global fitting of the data for WT NS3 (triangles) and the NS3ΔC7 (squares) construct with equation 3 as described in Materials and Methods. Best fits for WT data yielded n = 2.7 ± 0.11 and k = 0.063 ± 0.005 min−1. Best fits for NS3ΔC7 data yielded n = 3.5 ± 0.15 and k = 0.22 ± 0.01 min−1. For definitions of n and k, see the legend to Fig. 6.
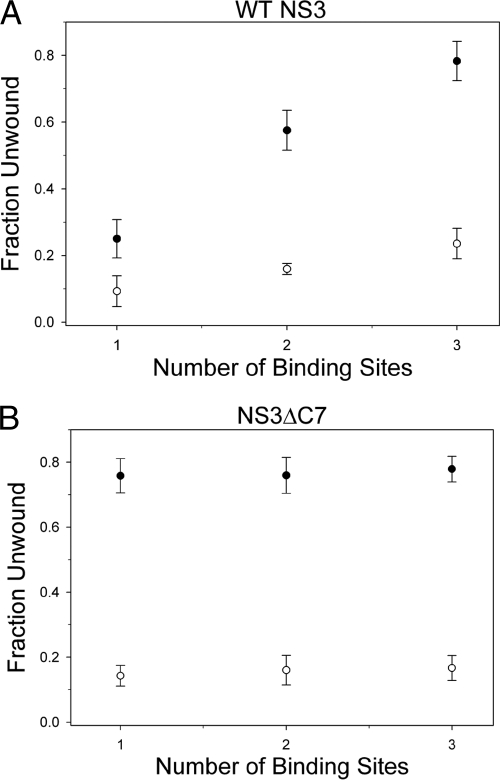
A NS3ΔC7 monomer unwinds the RNA duplex.
One way to explore the functional oligomeric state of a helicase is to examine the effects of the substrate overhang length (42). Based on the binding site size of NS3 determined by structural and biochemical methods (14, 22, 23, 42), we designed a family of RNA substrates with a fixed duplex length that is flanked by various single-stranded 3′-overhang lengths that can accommodate one, two, or three NS3 molecules (6-nt, 14-nt, and 20-nt overhangs, respectively). Qualitatively, the WT NS3 data reveal a strong positive correlation in the fraction of unwound substrate versus the overhang length (Fig. 8A). These results indicate that the number of WT NS3 molecules bound to the overhang strand contributes in an additive manner to the final unwinding amplitude. Fitting the data reveals that each additional binding site increases the unwinding amplitude by 7% ± 0.2% (equation 4; see Materials and Methods). A similar correlation in overhang length and unwinding amplitude had been reported in previous studies using the NS3hel construct (described as the “functional interaction model” between helicase molecules) (23). However, due to the different nucleic acid substrates and NS3 protein constructs used, we cannot make a quantitative comparison between the two studies.
Fig. 8.
An NS3ΔC7 monomer unwinds the RNA duplex. Functional complex formation of WT NS3 (A) or NS3ΔC7 (B) was measured on 34-bp RNA substrates containing a 3′-overhang length of 6, 14, or 20 nt, which accommodate 1, 2, or 3 helicase molecules, respectively. The trap was either a 60-nt DNA noncomplementary trap (open circles) or a 34-nt RNA complementary trap (closed circles). Graphs indicate the final, corrected fraction of RNA duplex unwound versus the number of binding sites on the 3′ overhang. Error bars represent the standard deviations determined from at least three, independent experiments.
The data for NS3ΔC7 did not display any correlation between overhang length and amplitude, within experimental uncertainty (Fig. 8B), suggesting that the unwinding amplitude is independent of overhang length. This finding indicates that an NS3ΔC7 monomer is likely to unwind each RNA duplex. It also suggests that NS3ΔC7 is incapable of forming functional interactions between protein molecules even when presented with the opportunity (such as long overhangs that stimulate formation of functional interactions between WT NS3 molecules) (23, 42). Thus, the altered conformation of NS3ΔC7, or the lack of a C-terminal tail itself, appears to inhibit formation of functional interactions between NS3ΔC7 molecules.
As a secondary and independent method to confirm these findings, we changed the chemical and nucleotide composition of the helicase trap. The trap used thus far is an unstructured, single-stranded DNA polymer that bears no sequence identity to either strand of the duplex substrate. Changing the trap to a single-stranded RNA polymer of identical sequence and length to the displaced strand of the duplex substrate increases the observed unwinding amplitude without affecting the rate constant for functional complex formation (6, 42).
Using a 34-nt RNA trap, we observed a direct correlation in the WT NS3 data similar to the data when using a DNA trap (Fig. 8A). However, the amplitudes of NS3ΔC7 unwinding were independent of the number of protein binding sites (Fig. 8B). The fitting of the WT NS3 data revealed that each increase in the overhang length increased the final unwinding amplitude by 27% ± 3%. These results validate our initial findings using the DNA trap and support our conclusion that an NS3ΔC7 monomer unwinds the RNA duplex.
DISCUSSION
Structural and functional effects of the β-interaction.
Here, we report that the presence or absence of a β-sheet interaction between protease and helicase domains of NS3 changes the structural and functional attributes of the protein (Fig. 1). When the β-interaction is removed, limited proteolysis experiments indicate that the C terminus of NS3ΔC7 becomes accessible to solvent (Fig. 2). In the absence of the β-interaction, it is unlikely that the helicase and protease domains of NS3ΔC7 maintain a compact conformation because the domains do not have a sufficiently extensive interaction interface. The interface between the helicase and protease domains of WT NS3 results in a buried surface area of ∼900 Å2 (53). Of this total area, which is apparent from the crystal structure, the β-interaction contributes ∼500 Å2 and is stabilized by an extensive hydrogen bonding network between the two domains. The remaining area of ∼400 Å2 is provided by interactions involving the flexible linker region that covalently connects the protease and helicase domains (Fig. 1), and it is not particularly hydrophobic. Because specific protein interfaces typically involve buried surface areas of 1,600 ± 400 Å2 (50) and have a high concentration of nonpolar residues (35), the remaining ∼400 Å2 is too small to represent a stable interaction interface. Therefore, we expect the protease domain of NS3ΔC7 to move freely and sample different conformational states, as observed for other Flaviviridae NS3 proteins (4, 27, 28).
Hydrodynamic measurements obtained using ultracentrifugation sedimentation velocity experiments provide direct evidence that WT NS3 and NS3ΔC7 adopt globally different shapes in solution (Fig. 3). A lower sedimentation coefficient argues that NS3ΔC7 adopts an extended conformation compared to WT NS3, and the broadening of the NS3ΔC7 sedimentation peak points to the presence of multiple extended conformations. Notably, DENV NS3 crystallized in two different extended conformations in which the protease domain had rotated ∼161° with respect to the helicase domain (27, 28). Therefore, the distribution of conformations that we observe during sedimentation velocity experiments on NS3ΔC7 are likely to derive from the protease domain as it samples different conformations in the extended, open state. Taken together, our structural analysis and hydrodynamic studies show that NS3ΔC7 adopts an extended conformation that is likely to resemble the extended conformations of other Flaviviridae NS3 proteins in which the protease domain sits beneath the ATP binding site of the helicase domain (4, 27, 28). The final orientation of the protein on a lipid bilayer is shown in Fig. S3 in the supplemental materrial.
The different conformations of WT NS3 and NS3ΔC7 result in different levels of functional activities. Specifically, we observe that NS3ΔC7 forms functional complexes with unwinding substrates faster than WT NS3 (Fig. 4). WT NS3 requires long incubation times to reach its maximal activity because the β-interaction locks it in an inactive, compact conformation. A slow isomerization step is required (reflected by k1; scheme 1) before WT NS3 can form functional unwinding complexes with RNA. Disruption of the β-interaction and adoption of an extended conformation are obligatory steps that precede formation of a productive complex with the unwinding substrate (a step reflected by k2). Already in the extended conformation, NS3ΔC7 bypasses the need for an isomerization step and quickly proceeds to form functional complexes and unwind the substrate. This behavior is most evident under reaction conditions that maintain WT NS3 in the locked, compact conformation by stabilizing the β-interaction (e.g., 15% glycerol or 150 mM NaCl) (Fig. 6 and 7). Collectively, these findings indicate that the extended conformation of NS3ΔC7 is poised to engage its target substrates while the compact conformation of WT NS3 is not.
Implications for viral replication.
Numerous in vitro studies indicate that the NS3 helicase has multiple activities, and it is possible that these activities reflect its diverse roles in vivo. The various activities attributed to NS3 include unwinding of duplex substrates, displacement of proteins bound to nucleic acids, translocation along single-stranded nucleic acids, and packaging of the RNA genome to form infectious viral particles (19, 30, 31). To initiate any of these tasks, NS3 must form productive interactions with its target substrate. Here we show that, for helicase activity, functional complex formation requires an extended conformation, and this form of NS3 is required for productive engagement with RNA duplex substrates. Consistent with previous studies in which replication-competent forms of NS3 were modeled onto lipid bilayers, we propose that the extended conformation is the biologically relevant form of NS3 for helicase activity (9, 27).
Why is it necessary for NS3 to undergo such a large conformational change? The isomerization described here would allow a multifunctional enzyme such as NS3 to coordinate the different enzymatic activities of its two functional domains. After the protease domain performs the peptide cleavage events that are required for releasing downstream viral proteins from the precursor polyprotein, the mature viral proteins can then assemble into a multiprotein complex that replicates the viral genome. During this process, NS3 must switch from its role as a protease and begin to operate exclusively as a helicase. It is possible that the conformational change described here is the key structural event that facilitates this switch in function.
Nonetheless, the C-terminal β-strand clearly has an innate affinity for the protease domain, and its presence in WT NS3 may cause the protein to alternate between compact and extended conformations even during the course of RNA binding and unwinding. It is probably important for NS3 to prevent the β-interaction from forming while it functions as a helicase. In vivo, NS3 is docked within a large multiprotein complex, and the extended conformation is probably maintained through protein-protein interactions. In their natural context, helicases rarely function in isolation, and they are usually components of larger complexes that contain cofactor proteins for regulating helicase function (25, 45). The sites of intermolecular interaction between NS3 and other proteins of the HCV replication complex may therefore serve as attractive drug targets for inhibiting viral replication.
Supplementary Material
ACKNOWLEDGMENTS
We thank Meigang Gu (Rockefeller) for early discussions of the project; Arnon Henn, Wenxiang Cao, Michael Bradley, and Enrique de la Cruz (Yale) for valuable discussions on fitting of the data; Olga Fedorova for synthesis of RNA substrates; and Dahai Luo, Christian Matranga, Victor Serebrov, and other members of the Pyle lab for reading and providing critical comments on the manuscript. We thank the Yale Chemical and Biophysical Instrumentation Center for use of the analytical ultracentrifuge.
This work was supported by NIH training grants T32GM008283 (S.C.D.) and T32GM007223 (A.S.K.). A.M.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 16 February 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Ali J. A., Lohman T. M. 1997. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275:377–380 [DOI] [PubMed] [Google Scholar]
- 2. Appel N., Schaller T., Penin F., Bartenschlager R. 2006. From structure to function: new insights into hepatitis C virus RNA replication. J. Biol. Chem. 281:9833–9836 [DOI] [PubMed] [Google Scholar]
- 3. Arakawa T., Timasheff S. N. 1985. Theory of protein solubility. Methods Enzymol. 114:49–77 [DOI] [PubMed] [Google Scholar]
- 4. Assenberg R., et al. 2009. Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J. Virol. 83:12895–12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belon C. A., Frick D. N. 2009. Helicase inhibitors as specifically targeted antiviral therapy for hepatitis C. Future Virol. 4:277–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beran R. K., Bruno M. M., Bowers H. A., Jankowsky E., Pyle A. M. 2006. Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J. Mol. Biol. 358:974–982 [DOI] [PubMed] [Google Scholar]
- 7. Beran R. K., Pyle A. M. 2008. Hepatitis C viral NS3-4A protease activity is enhanced by the NS3 helicase. J. Biol. Chem. 283:29929–29937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beran R. K., Serebrov V., Pyle A. M. 2007. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J. Biol. Chem. 282:34913–34920 [DOI] [PubMed] [Google Scholar]
- 9. Brass V., et al. 2008. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc. Natl. Acad. Sci. U. S. A. 105:14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dashnau J. L., Nucci N. V., Sharp K. A., Vanderkooi J. M. 2006. Hydrogen bonding and the cryoprotective properties of glycerol/water mixtures. J. Phys. Chem. B 110:13670–13677 [DOI] [PubMed] [Google Scholar]
- 11. Dumont S., et al. 2006. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439:105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gill S. C., von Hippel P. H. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319–326 [DOI] [PubMed] [Google Scholar]
- 13. Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gu M., Rice C. M. 2010. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. U. S. A. 107:521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jankowsky E., Gross C. H., Shuman S., Pyle A. M. 2000. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature 403:447–451 [DOI] [PubMed] [Google Scholar]
- 16. Jennings T. A., et al. 2008. RNA unwinding activity of the hepatitis C virus NS3 helicase is modulated by the NS5B polymerase. Biochemistry 47:1126–1135 [DOI] [PubMed] [Google Scholar]
- 17. Kawai T., et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 18. Kawaoka J., Jankowsky E., Pyle A. M. 2004. Backbone tracking by the SF2 helicase NPH-II. Nat. Struct. Mol. Biol. 11:526–530 [DOI] [PubMed] [Google Scholar]
- 19. Khaki A. R., et al. 2010. The macroscopic rate of nucleic acid translocation by hepatitis C virus helicase NS3h is dependent on both sugar and base moieties. J. Mol. Biol. 400:354–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim J. L., et al. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89–100 [DOI] [PubMed] [Google Scholar]
- 21. Lam A. M., Frick D. N. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 80:404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levin M. K., Gurjar M., Patel S. S. 2005. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat. Struct. Mol. Biol. 12:429–435 [DOI] [PubMed] [Google Scholar]
- 23. Levin M. K., Wang Y. H., Patel S. S. 2004. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 279:26005–26012 [DOI] [PubMed] [Google Scholar]
- 24. Li K., et al. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. U. S. A. 102:2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lohman T. M., Tomko E. J., Wu C. G. 2008. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 9:391–401 [DOI] [PubMed] [Google Scholar]
- 26. Lucius A. L., Maluf N. K., Fischer C. J., Lohman T. M. 2003. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 85:2224–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luo D., et al. 2010. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J. Biol. Chem. 285:18817–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo D., et al. 2008. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 82:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luo D., et al. 2008. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 27:3209–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma Y., Yates J., Liang Y., Lemon S. M., Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matlock D. L., et al. 2010. Investigation of translocation, DNA unwinding, and protein displacement by NS3h, the helicase domain from the hepatitis C virus helicase. Biochemistry 49:2097–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meylan E., et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172 [DOI] [PubMed] [Google Scholar]
- 33. Moradpour D., Penin F., Rice C. M. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 34. Myong S., Bruno M. M., Pyle A. M., Ha T. 2007. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science 317:513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nooren I. M., Thornton J. M. 2003. Structural characterisation and functional significance of transient protein-protein interactions. J. Mol. Biol. 325:991–1018 [DOI] [PubMed] [Google Scholar]
- 36. Pang P. S., Jankowsky E., Planet P. J., Pyle A. M. 2002. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 21:1168–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pyle A. M. 2008. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 37:317–336 [DOI] [PubMed] [Google Scholar]
- 38. Raibekas A. A., Massey V. 1997. Glycerol-assisted restorative adjustment of flavoenzyme conformation perturbed by site-directed mutagenesis. J. Biol. Chem. 272:22248–22252 [DOI] [PubMed] [Google Scholar]
- 39. Rajagopal V., Gurjar M., Levin M. K., Patel S. S. 2010. The protease domain increases the translocation stepping efficiency of the hepatitis C virus NS3-4A helicase. J. Biol. Chem. 285:17821–17832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raney K. D., Sharma S. D., Moustafa I. M., Cameron C. E. 2010. Hepatitis C virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J. Biol. Chem. 285:22725–22731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., Schubert D. 2002. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 82:1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Serebrov V., Beran R. K., Pyle A. M. 2009. Establishing a mechanistic basis for the large kinetic steps of the NS3 helicase. J. Biol. Chem. 284:2512–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Serebrov V., Pyle A. M. 2004. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature 430:476–480 [DOI] [PubMed] [Google Scholar]
- 44. Seth R. B., Sun L., Ea C. K., Chen Z. J. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 45. Silverman E., Edwalds-Gilbert G., Lin R. J. 2003. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene 312:1–16 [DOI] [PubMed] [Google Scholar]
- 46. Tai C. L., Chi W. K., Chen D. S., Hwang L. H. 1996. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol. 70:8477–8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Timasheff S. N. 1993. The control of protein stability and association by weak interactions with water: how do solvents affect these processes? Annu. Rev. Biophys. Biomol. Struct. 22:67–97 [DOI] [PubMed] [Google Scholar]
- 48. Wincott F., et al. 1995. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 23:2677–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Winder J. S., Walker J. M. 1993. Carboxypeptidase Y (EC 3.4.16.1). Methods Mol. Biol. 16:313–318 [DOI] [PubMed] [Google Scholar]
- 50. Wodak S. J., Janin J. 2002. Structural basis of macromolecular recognition. Adv. Protein Chem. 61:9–73 [DOI] [PubMed] [Google Scholar]
- 51. Xu L. G., et al. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727–740 [DOI] [PubMed] [Google Scholar]
- 52. Yao N., et al. 1997. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 4:463–467 [DOI] [PubMed] [Google Scholar]
- 53. Yao N., Reichert P., Taremi S. S., Prosise W. W., Weber P. C. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure 7:1353–1363 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.