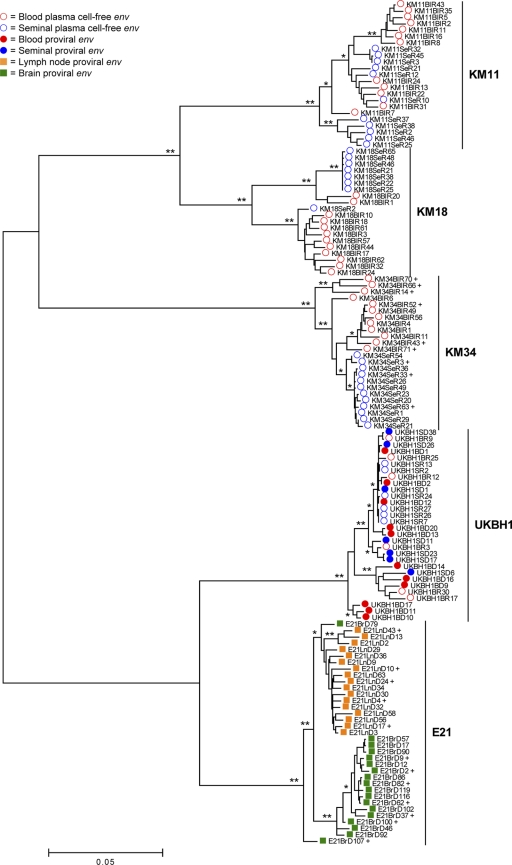
Fig. 1.
Phylogenetic assessment of env compartmentalization from immunologically discrete tissues. A midpoint-rooted ML phylogenetic reconstruction for the combined HIV-1 env alignment generated using the GTR+I+Γ4 substitution model (74, 88, 89) is shown. Branch lengths are in accordance with the scale bar and are proportional to genetic distance. Significance of clustering of isolates was assessed via bootstrap resampling of the sequence data derived from 1000 replications. *, Values >70%; **, values >99%. Isolates marked with a “+” were selected for subsequent phenotyping.