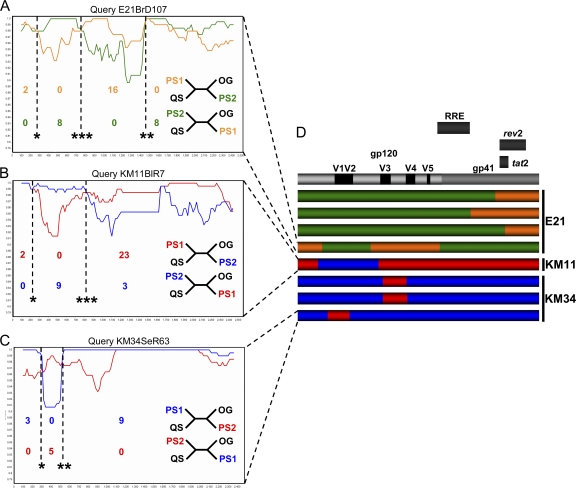
Fig. 2.
Evidence for intercompartment mosaicism. Example similarity plots and informative site arrays were used to characterize intercompartment mosaics. Plots were generated in SimPlot with a sliding window size of 200 bp and a step size of 20 bp and represent a query mosaic compared to two parental sequences. Dotted vertical lines indicate absolute breakpoint positions correlated with diversity plot crossover points. Associated P values (*, P < 0.05; **, P < 0.01; ***, P < 0.001) are derived from informative site arrays (described in Materials and Methods and displayed in Table 2). Relative numbers of informative sites shared by query sequences with parental strains are displayed below regions of differential homology. Four taxon trees consistent with these sites are displayed to the left: QS, query sequence; PS1, parental sequence 1; PS2, parental sequence 2; OG, outgroup. (A) Query mosaic E21BrD107 compared to parental sequences E21BrD90 and E21LnD56. (B) Query mosaic KM11BlR7 compared to parental sequences KM11BlR22 and KM11SeR2. (C) Query mosaic KM34SeR63 compared to parental sequences KM34BlR11 and KM34SeR26. (D) Schematic diagram depicting breakpoint locations in eight characterized mosaics, displayed relative to the positions of functionally defined regions in the HXB2 reference env located above. The patient from which each mosaic was isolated is detailed to the right of each individual recombinant env schematic. Regions of differential homology are color coded.