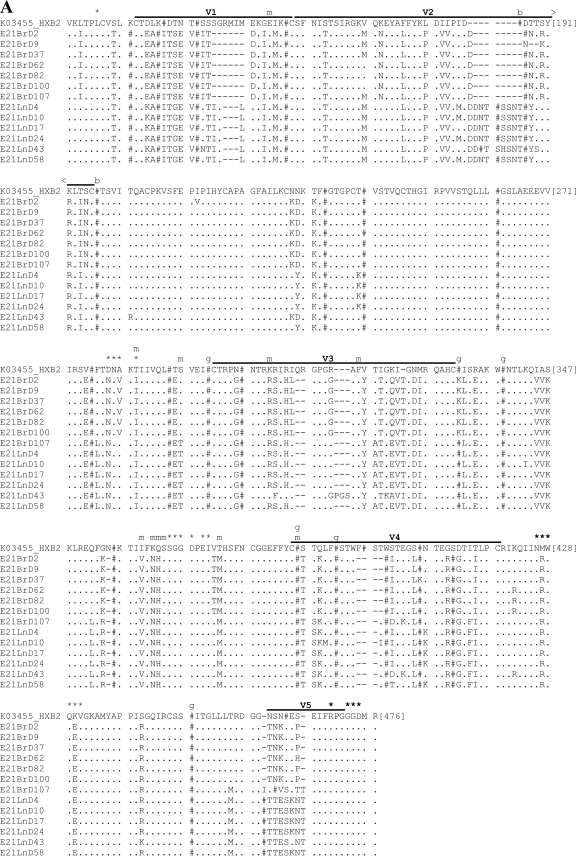
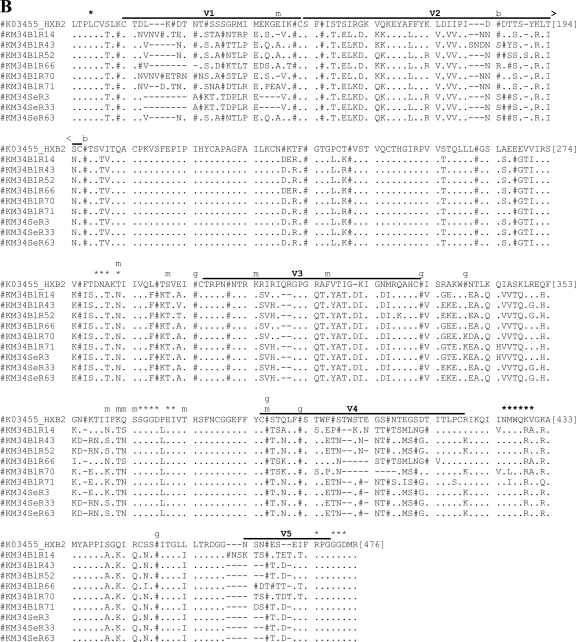
Fig. 5.
Amino acid sequence alignments for functional gp160s. Functional gp160s from patient E21 (A) and patient KM34 (B) are aligned relative to the HXB2 reference strain, and all residue numbering is relative to homologous positions in HXB2 gp160. Within the alignment, asparagine (N) residues that are likely to be glycosylated have been replaced by “#”. Horizontal lines located above alignment segments indicate the position of the variable regions V1 to V5. Symbols or letters located directly above alignment segments highlight the functionally important residues: *, CD4 contact residue; m, additional residue previously implicated in modulation of macrophage tropism; b, residue previously implicated in modulating sensitivity to b12; and g, residue previously implicated in modulating sensitivity to 2G12.