Abstract
Carriage of the natural killer (NK) receptor genotype KIR3DL1*h/*y with its HLA-B*57 ligand (*h/*y+B*57) is associated with slow time to AIDS and low viral load (VL). To provide a functional basis for these epidemiological observations, we assessed whether HIV-1-infected slow progressors (SP) carrying the *h/*y+B*57 compound genotype would have increased NK cell polyfunctional potential in comparison to SP with other killer immunoglobulin-like receptor (KIR)/HLA compound genotypes and whether this enhanced polyfunctionality was dependent upon the coexpression of both KIR3DL1*h/*y and HLA-B*57. The functional potential of NK cells was investigated by stimulating peripheral blood mononuclear cells with HLA-devoid targets or single HLA transfectants. Multiparametric flow cytometry was used to detect NK cells with seven functional profiles representing all permutations of CD107a expression and gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α) secretion. NK cells from individuals carrying KIR3DL1 receptor–HLA-Bw4 ligand pairs had greater trifunctional responses than those from KIR3DL1 homozygotes (hmz), who were Bw6 homozygotes. NK cells from subjects carrying the *h/*y+B*57 genotypes exhibited the highest trifunctional potential, and this was dependent on cocarriage of the NK receptor and its ligand. Trifunctional cells secreted more of each function tested on a per-cell basis than each corresponding monofunctional NK subset. Although VL influenced NK functionality, individuals with defined KIR/HLA genotypes exhibited differences in NK cell polyfunctionality that could not be accounted for by VL alone. The protective effect of HLA-B*57 on slow progression to AIDS and low VL may be mediated through its interaction with KIR3DL1 alleles to educate NK cells for potent activity upon stimulation.
INTRODUCTION
Natural killer (NK) cells are key players in host innate immune defenses. They function in the early response to virus-infected and transformed cells without prior sensitization (4, 49). Through the release of preformed granules containing perforin and granzyme, NK cells can directly lyse tumor cells and virally infected targets. In addition, they can secrete large amounts of proinflammatory cytokines and chemokines, which promote interactions with dendritic cells (DC), monocytes, and granulocytes and influence the evolution of the adaptive immune response (17, 29, 47).
NK function is tightly regulated by the integration of inhibitory and activation signals transmitted through germ line-encoded cell surface receptors, which include alleles of the killer immunoglobulin-like receptor (KIR) region genes. KIRs are expressed on NK cells and some T-cell subsets (29). The most polymorphic locus among KIR region genes is KIR3DL1, which carries both inhibitory KIR3DL1 (henceforth 3DL1) and activating KIR3DS1 (henceforth 3DS1) alleles (44). 3DL1 alleles can be further classified according to their expression levels on the cell surface into high (*h), low/intermediate (*l), and null (*004) unexpressed alleles. (20, 44, 46, 52). Genotypes homozygous for 3DL1 can be divided into two groups: *h/*y (where *y can be either another *h allele or *004) with no *l alleles and *l/*x (where *x can be either another *h, *l, or *004) with at least one *l allele (36). Cell surface staining with DX9, a monoclonal antibody (MAb) specific for 3DL1, shows that 3DL1 levels are higher on NK cells from carriers of the *h/*y genotype than on those from carriers of the *l/*x genotype (52).
The ligands for 3DL1 receptors belong to the Bw4 group of major histocompatibility complex (MHC) class I B alleles that are distinguished from the Bw6 subset by amino acids present between positions 77 and 83 of the HLA heavy chain (22, 33, 50). Bw4 alleles can be further divided into those with an isoleucine at amino acid 80 (Bw4*80I) or a threonine at this position (Bw4*80T) of the HLA heavy chain (13). Bw4*80I alleles are the preferred ligands for 3DL1 receptors (18, 27).
Epidemiological studies have shown an association between certain KIR/HLA combined genotypes with time to AIDS and viral load (VL) set point (35, 36). Compared to Bw6 homozygotes (hmz) with no alleles that interact with KIR, the 3DL1/Bw4 receptor-ligand combination, which has the most potent effect on favorable HIV disease outcomes, is the one that combines 3DL1*h/*y with HLA-B*57, (*h/*y+B*57) (36). NK cells from HIV-uninfected persons carrying the *h/*y+B*57 genotype respond to stimulation with the HLA-devoid K562 cell line with a higher frequency of functional cells exhibiting multiple functions than NK cells from Bw6 hmz (9).
B*57 is a protective allele in the context of HIV infection and has a higher frequency in HIV-infected slow progressors (SP) than in HIV disease progressors or healthy controls (19, 26, 42). Part of its effect on slow time to AIDS and VL set point is mediated through the adaptive arm of the immune response via CD8+ T cells that recognize B*57 restricted epitopes (3, 21, 30, 42, 43). However, because B*57 is a Bw4*80I allele and therefore a ligand for 3DL1 receptors, it has the capacity to participate in the MHC class I-dependent NK cell functional maturation process termed “education,” or “licensing,” that renders NK cells functionally competent (54).
This study investigated the relationship between NK cell functional potential and KIR/HLA genotype in a population of HIV-infected SP. We found significant differences in the percentages of contributions of NK cells with three functions (secretion of gamma interferon [IFN-γ] and tumor necrosis factor alpha [TNF-α] and expression of CD107a) following stimulation with K562 when from 3DL1 hmz who carried at least one copy of a Bw4 allele versus a Bw6 hmz genotype. Of those who carried the 3DL1 and Bw4 NK receptor-ligand combination, it was the carriers of the *h/*y+B*57 genotype who exhibited the highest NK functional potential. Expression of neither 3DL1*h/*y nor B*57 alone was sufficient to support the level of stimulated trifunctionality observed in NK cells from *h/*y+B*57 carriers. Each of the functions exhibited by trifunctional NK cells was more potent on a per cell basis than the same function in the monofunctional NK subsets. We used a panel of transfectants expressing single HLA alleles to further confirm that disruption of 3DL1 signaling was responsible for elevated functional levels in NK cells from *h/*y+B*57 carriers stimulated with HLA-devoid cells. Transfectants expressing the Bw4*80I alleles B*57:01 and B*27:02 suppressed the function of NK cells from *h/*y+B*57 carriers, while the transfectant expressing the Bw6 B*35:02 allele did not.
MATERIALS AND METHODS
Study population.
The study population included 57 HIV-infected SP. Thirty-six SP were recruited from the Canadian Cohort of HIV Infected Slow Progressors, and 21 were from a cohort followed at the National Institute of Allergy and Infectious Diseases (NIAID) (41). The term “SP” was used here to define treatment-naïve HIV-infected subjects who maintained absolute CD4 counts above 400 cells/mm3 for more than 7 years, elite controllers (EC) monitored for at least 1 year with a VL of <50 copies/ml of plasma, and viral controllers (VC) monitored for at least 1 year with a VL of <3,000 copies/ml of plasma. Information on CD4 and CD8 T-cell counts, VL and duration of infection at time of testing, 3DL1 genotype, and HLA type of the study population is provided in Table 1. All participants were 3DL1 hmz. This allowed them to be classified as 3DL1*h/*y or *l/*x and eliminated the possible confounding influence of an activating 3DS1 allele on NK functional potential (8, 36, 52). Informed consent was obtained from all study participants, and research adhered to the ethical guidelines of the authors' institutions. For certain comparisons, we also included results generated from 11 HIV-1 negative *h/*y+B*57 carriers whose characteristics have been previously described (9) and 12 HIV-1-infected untreated viremic progressors recruited from the Montreal Primary Infection Cohort. The descriptive characteristics of these 12 HIV-infected viremic progressors are included in Table 1.
Table 1.
Study population characteristics
| Patient no. | Log10 viral load | T-cell count (cells/mm3) |
Duration of infection (yr) | Age (yr) | Gender | Ethnicity | HLA-B type |
KIR3DL1 allotype group | ||
|---|---|---|---|---|---|---|---|---|---|---|
| CD4 | CD8 | Allele 1 | Allele 2 | |||||||
| 1 | 3.54 | 440.0 | 350.0 | 14 | 44 | Male | Caucasian | B*07:02 | B*51:01 | *h/*y |
| 2 | 3.15 | 420.0 | 1,180.0 | 15 | 44 | Female | Caucasian | B*44:02 | B*52:01 | *h/*y |
| 3 | 3.70 | 682.0 | 1,643.0 | 1 | 53 | Male | Caucasian | B*27:01 | B*51:01 | *h/*y |
| 4 | 3.91 | 830.0 | 1,700.0 | 20 | 60 | Male | Caucasian | B*27:03 | B*57:01 | *h/*y |
| 5 | 3.90 | 770.0 | 990.0 | 10 | 61 | Male | Caucasian | B*40:02 | B*57:01 | *h/*y |
| 6 | 4.12 | 788.0 | 2,498.0 | 10 | 34 | Male | Caucasian | B*15:01 | B*27:03 | *h/*y |
| 7 | 1.70 | 1,040.0 | 1,090.0 | 3 | 37 | Male | Caucasian | B*27:05 | B*40:02 | *h/*y |
| 8 | 1.70 | 580.0 | 1,440.0 | 8 | 35 | Male | Caucasian | B*39:01 | B*51:01 | *h/*y |
| 9 | 2.43 | 535.0 | 793.0 | 1 | 35 | Female | African | B*49:01 | B*53:01 | *h/*y |
| 10 | 3.53 | 440.0 | 750.0 | 1 | 47 | Male | Caucasian | B*27:03 | B*57:01 | *h/*y |
| 11 | 1.70 | 343.0 | 804.0 | 10 | 26 | Male | Caucasian | B*14:02 | B*15:01 | *h/*y |
| 12 | 1.70 | 489.0 | 672.0 | 1 | 31 | Female | African | B*27:03 | B*57:01 | *l/*x |
| 13 | 3.38 | 470.0 | 600.0 | 7 | 38 | Male | Caucasian | B*07:02 | B*57:01 | *l/*x |
| 14 | 2.35 | 580.0 | 860.0 | 13 | 53 | Male | Asian | B*13:01 | B*57:01 | *l/*x |
| 15 | 3.73 | 590.0 | 1,650.0 | 6 | 50 | Female | African | B*15:03 | B*44:02 | *h/*y |
| 16 | 2.87 | 1,262.0 | 731.0 | 2 | 39 | Female | Caucasian | B*35:01 | B*57:01 | *h/*y |
| 17 | 1.70 | 410.0 | 560.0 | 2 | 24 | Male | Caucasian | B*57:01 | B*73:01 | *h/*y |
| 18 | 1.70 | 870.0 | 550.0 | 1 | 46 | Male | African | B*15:01 | B*44:02 | *l/*x |
| 19 | 1.70 | 715.0 | 384.0 | 5 | 32 | Female | African | B*14:02 | B*44:03 | *l/*x |
| 20 | 1.70 | 740.0 | 1,180.0 | 4 | 70 | Male | African | B*08:01 | B*15:01 | *l/*x |
| 21 | 2.80 | 360.0 | 1,610.0 | 3 | 37 | Male | Caucasian | B*35:01 | B*58 | *l/*x |
| 22 | 1.70 | 720.0 | 830.0 | 14 | 47 | Female | Caucasian | B*07:02 | B*38:01 | *l/*x |
| 23 | 1.70 | 396.0 | 998.0 | 3 | 38 | Male | Caucasian | B*15:01 | B*27:01 | *l/*x |
| 24 | 2.81 | 500.0 | 580.0 | 1 | 33 | Male | Caucasian | B*44:03 | B*57:01 | *l/*x |
| 25 | 3.40 | 700.0 | 920.0 | 16 | 48 | Male | Caucasian | B*44:03 | B*57:01 | *l/*x |
| 26 | 3.24 | 500.0 | 530.0 | 6 | 35 | Male | Asian | B*15:02 | B*51:02 | *l/*x |
| 27 | 1.70 | 1,200.0 | 860.0 | 12 | 41 | Male | Caucasian | B*40:01 | B*57:01 | *h/*y |
| 28 | 3.54 | 400.0 | 530.0 | 16 | 39 | Female | African | B*49:01 | B*58:02 | *h/*y |
| 29 | 3.76 | 1,487.0 | 712.0 | 13 | 40 | Female | Caucasian | B*07:02 | B*13:01 | *h/*y |
| 30 | 4.21 | 650.0 | 1,460.0 | 6 | 38 | Male | Caucasian | B*07:02 | B*57:01 | *h/*y |
| 31 | 1.70 | 1,239.0 | 1,267.0 | 18 | 58 | Male | African-American | B*57 | B*44 | *h/*y |
| 32 | 1.70 | 378.0 | 618.0 | 22 | 50 | Female | Caucasian | B*57 | B*27 | *l/*x |
| 33 | 1.70 | 1,048.0 | 1,556.0 | 10 | 49 | Male | Caucasian | B*57 | B*13 | *h/*y |
| 34 | 1.70 | 1,443.0 | 895.0 | 3 | 40 | Female | African-American | B*57 | B*07 | *h/*y |
| 35 | 1.70 | 371.0 | 436.0 | 24 | 59 | Female | Caucasian | B*57 | B*57 | *h/*y |
| 36 | 1.70 | 664.0 | 1,120.0 | 21 | 58 | Male | Caucasian | B*57 | B*44 | *l/*x |
| 37 | 1.70 | 833.0 | 590.0 | 11 | 54 | Male | African-American | B*57 | B*15 | *h/*y |
| 38 | 3.21 | 803 | 1,109 | 19 | 59 | Female | African-American | B*57 | B*81 | *l/*x |
| 39 | 1.70 | 1,106.0 | 1,240.0 | 15 | 44 | Male | Caucasian | B*57 | B*08 | *l/*x |
| 40 | 1.70 | 1,362.0 | 1,055.0 | 21 | 46 | Male | African-American | B*57 | B*81 | *h/*y |
| 41 | 1.70 | 684.0 | 566.0 | 7 | 38 | Male | African-American | B*57 | B*42 | *h/*y |
| 42 | 1.70 | 502 | 477 | 21 | 50 | Male | Caucasian | B*57 | B*37 | *h/*y |
| 43 | 2.06 | 718.0 | 1,637.0 | 9 | 52 | Male | African-American | B*57 | B*53 | *h/*y |
| 44 | 1.70 | 1,613.0 | 1,428.0 | 7 | 50 | Female | African-American | B*57 | B*53 | *h/*y |
| 45 | 1.70 | 807.0 | 1,084.0 | 11 | 43 | Male | Caucasian | B*57 | B*51 | *h/*y |
| 46 | 1.70 | 499.0 | 202.0 | 8 | 53 | Female | African-American | B*57 | B*42 | *h/*y |
| 47 | 3.17 | 568.0 | 408.0 | 20 | 56 | Male | African-American | B*57 | B*57 | *h/*y |
| 48 | 1.70 | 485.0 | 277.0 | 20 | 47 | Female | African-American | B*57 | B*15 | *h/*y |
| 49 | 3.22 | 708.0 | 1,816.0 | 7 | 39 | Male | Caucasian | B*57 | B*44 | *h/*y |
| 50 | 1.70 | 500.0 | 1,284.0 | 14 | 40 | Male | African-American | B*57 | B*81 | *h/*y |
| 51 | 1.70 | 801 | 1,012 | 18 | 56 | Male | Caucasian | B*57 | B*07 | *h/*y |
| 52 | 4.41 | 650.0 | 2,110.0 | 17 | 62 | Male | African | B*38:02 | B*51:01 | *h/*y |
| 53 | 1.70 | 636.0 | 663.0 | 3 | 57 | Female | African | B*07:02 | B*42 | *l/*x |
| 54 | 3.30 | 447.0 | 455.0 | 2 | 53 | Male | Caucasian | B*07:02 | B*14:02 | *h/*y |
| 55 | 5.21 | 546.0 | 572.0 | 17 | 47 | Male | Caucasian | B*39:01 | B*35:01 | *h/*y |
| 56 | 3.30 | 985.0 | 605.0 | 3 | 39 | Male | Caucasian | B*07:02 | B*07:02 | *h/*y |
| 57 | 3.41 | 350.0 | 730.0 | 4 | 40 | Male | Caucasian | B*14:02 | B*14:02 | *h/*y |
| 58 | 3.62 | 370.0 | 1,090.0 | 2 | 23 | Male | Caucasian | B*15:01 | B*15:01 | *h/*y |
| 59 | 3.40 | 260.0 | 550.0 | 2 | 41 | Male | Caucasian | B*14:02 | B*18:01 | *l/*x |
| 60 | 3.35 | 490.0 | 710.0 | 2 | 28 | Male | Caucasian | B*39:05 | B*39:06 | *h/*y |
| 61 | 3.24 | 480.0 | 1,140.0 | 3 | 35 | Male | Caucasian | B*14:02 | B*14:02 | *h/*y |
| 62 | 3.90 | 430.0 | 430.0 | 1 | 43 | Male | Caucasian | B*07:19 | B*40:02 | *h/*y |
| 63 | 4.30 | 613.0 | 993.0 | 2 | 46 | Male | Caucasian | B*07:02 | B*15:01 | *h/*y |
| 64 | 4.33 | 300.0 | 460.0 | 1 | 46 | Male | Caucasian | B*39:01 | B*07:02 | *h/*y |
| 65 | 3.60 | 729.0 | 957.0 | 1 | 31 | Male | Caucasian | B*35:01 | B*57 | *h/*y |
| 66 | 3.71 | 686.0 | 1,498.0 | 10 | 60 | Male | African-American | B*45 | *57 | *h/*y |
| 67 | 3.72 | 1,114.0 | 896.0 | 20 | 56 | Female | African-American | B*81:01 | *57 | *h/*y |
| 68 | 4.24 | 900.0 | 1,200.0 | 1 | 32 | Male | Caucasian | B*44:02 | B*57:01 | *h/*y |
| 69 | 4.80 | 432.0 | 814.0 | 3 | 42 | Male | Caucasian | B*07:02 | B*57:01 | *h/*y |
MHC and KIR typing.
All subjects were typed for MHC class I alleles by sequence-based typing using kits from Atria Genetics (South San Francisco, CA) and using Assign software to interpret sequence information for allele typing (Conexio Genetics, Perth, Australia) as previously described (9). HLA-B alleles were classified as either Bw4 or Bw6 and Bw4 alleles as either Bw4*80I or Bw4*80T, depending on whether they had an isoleucine or threonine at position 80 of the HLA heavy chain. Bw6 hmz served as controls for the effect of NK education signals through 3DL1 on NK functional potential as Bw6 alleles do not interact with 3DL1 (13, 22)
3DL1/S1 genotyping was performed using two sets of primers specific for the 3DL1 and 3DS1 loci as previously described (8). Subjects were subsequently 3DL1 allotyped by identification of single nucleotide polymorphisms (SNP) corresponding to high-frequency 3DL1 alleles as previously described (9). In our study, we categorized 3DL1*005, *006, *007, *053, and *054 as *l alleles; *001, *002, *008, *009, *015, and *020 as *h alleles; and *004 as a null allele.
Cells.
Peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation (Ficoll-Paque; Pharmacia Uppsala, Sweden) from whole blood obtained by venipuncture into tubes containing EDTA anticoagulant or by leukapheresis as previously described (7). Cells were cryopreserved in 10% dimethyl sulfoxide (DMSO) (Sigma-Aldrich, St. Louis, MO) with 90% fetal bovine serum (FBS) (Wisent, St. Bruno, Quebec, Canada).
Assessment of NK cell functional potential. (i) NK cell activation.
Cryopreserved PBMC were thawed and resuspended at 106 cells/ml in RPMI 1640 that contained 10% FBS, 2 mM l-glutamine, 50 IU penicillin, and 50 μg/ml streptomycin (all from Wisent). Brefeldin A (5 μg/ml; Sigma-Aldrich), monensin (6 μg/ml [Golgi-Stop; BD Biosciences, Mississauga, Ontario, Canada]), and anti-CD107a-fluorescein isothiocyanate (FITC) monoclonal antibody (MAb) (BD Biosciences) were added to the cells.
PBMC (106 per condition) from 47 SP were stimulated with HLA-devoid K562 cells (American Type Culture Collection, Manassas, VA). PBMC (106 per condition) from 10 of the SP carrying *h/*y+B*57 were stimulated with the HLA-devoid Epstein-Barr virus (EBV)-transformed cell line 721.221 (hereafter, 221) or 221 transfectants expressing single HLA alleles (221-B*57:01, 221-B*27:02, and 221-B*35:02). For all stimulations, the PBMC/target ratio was 5:1 for 6 h at 37°C in a humidified 5% CO2 incubator. Included in all experiments were conditions in which PBMC were stimulated with medium alone as a negative control and with 1.25 μg/ml phorbol 12-myristate 13-acetate (PMA)–0.25 μg/ml ionomycin (Sigma Aldrich) as a positive control. Only data from cells that responded to the PMA-ionomycin positive control stimulus were included in analyses.
(ii) NK cell staining for phenotype and function.
After stimulation, cells were stained for viability using the Aqua LIVE/DEAD fixable dead cell stain kit (Invitrogen, Burlington, Ontario, Canada) following the manufacturer's instructions. Live PBMC were then stained for cell surface markers with anti-CD56-allophycocyanin (APC), anti-CD16-Pacific Blue (BD Biosciences), anti-CD3-phycoerythrin Texas Red conjugate (energy-coupled dye) (anti-CD3-ECD), and CD158e-phycoerythrin (PE) (i.e., Z27-PE; Beckman Coulter, Mississauga, Ontario, Canada) for 30 min. After being washed with phosphate-buffered saline (PBS) containing 1% FBS (Wisent) and 0.1% sodium azide (Sigma Aldrich), cells were fixed and permeabilized with the Invitrogen Fix and Perm kit and stained for intracellular cytokines with anti-IFN-γ-Alexa 700 and anti-TNF-α-PE-Cy7 MAbs (BD Biosciences). Cells were washed and fixed with a 1% paraformaldehyde solution (Fisher Scientific, Ottawa Ontario, Canada) and stored in the dark at 4°C until acquisition.
In parallel, 1 million unstimulated PBMCs were phenotyped for NK markers by staining for viability using the Aqua LIVE/DEAD fixable dead cell stain kit (Invitrogen) and for cell surface markers with anti-CD56-APC, anti-CD16-Pacific Blue (BD Biosciences), anti-CD3-ECD, and CD158e-PE (i.e., Z27-PE; Beckman Coulter, Mississauga, Ontario, Canada) for 30 min. After washing with PBS containing 1% FBS (Wisent) and 0.1% sodium azide (Sigma Aldrich), cells were washed and fixed with a 1% paraformaldehyde solution (Fisher Scientific) and stored in the dark at 4°C until acquisition.
Flow cytometry analysis.
Cells were acquired on an LSRII flow cytometer (BD Biosciences). Between 400,000 and 500,000 events were collected per sample. Data analysis was performed using FlowJo software version 8.7.1 (Tree Star, San Carlos, CA). The functional profiles of NK cells stimulated with K562, 221 and the 221 HLA transfectant panel were determined by the gating strategy shown in Fig. S1 in the supplemental material. NK cells were defined as CD3− CD56+/− CD16+/−. Boolean gating was used to identify seven NK cell functional profiles: i.e., trifunctional NK cells (CD107a+ IFN-γ+ TNF-α+), bifunctional NK cells (any combination of two of these functions), and monofunctional NK cells (any one of these functions). The Z27 antibody was used to assess the frequency and percent contribution of each of the seven possible NK cell functional profiles within the 3DL1+ or 3DL1− NK response. To obtain the frequency of each functional subset and then the percent contribution of each NK cell functional subset to the total K562-stimulated response, the negative control (medium alone) value for each of these functional subsets was subtracted from the corresponding K562-stimulated functional subset response to correct for background. To support analyses in which mean fluorescence intensities (MFI) of CD107a expression and IFN-γ and TNF-α were compared in NK cells having different functional profiles, Ultra Rainbow fluorescent particles (Spherotech, Cedarlane, Burlington, Ontario, Canada) were used to calibrate the LSRII flow cytometer. A total of 500,000 events were acquired for the MFI comparisons.
Generation of 221 cells expressing individual HLA alleles.
The cDNAs encoding HLA-B*57:01, HLA-B*27:02, and HLA-B*35:02 were amplified by PCR and cloned into a murine stem cell virus (MSCV)-based retroviral vector carrying the puromycin resistance gene under the control of an internal ribosome entry site (IRES). The full-length insert was sequenced, and the sequence was verified against the reference sequences in the IMGT/HLA database. HEK-293T cells were cotransfected with the retroviral construct and plasmids encoding Gag/Pol and vesicular stomatitis virus glycoprotein (VSV-G), and retrovirus-containing supernatant was harvested 48 and 96 h posttransfection. To generate 221 cells stably expressing individual HLA alleles, 105 221 cells were cultured in the retrovirus-containing supernatant for 3 days. Two days after this incubation, 1 μg/ml puromycin was added, and the cells were cultured until a stable polyclonal line was produced. The surface expression of the individual alleles was verified by flow cytometry, and following selection, all cells expressed the individual alleles.
Statistical analysis.
GraphPad Instat 3.05 and GraphPad Prism 4.01 were used for statistical analyses and graphical presentations. Mann-Whitney U tests were used for between-group comparisons of the percent contribution of trifunctional NK cells to the total NK cell response among SP. A Bonferonni correction was applied to situations in which multiple comparisons were made. The Spearman's correlation test was used to assess the significance of associations between the percent contribution of a response to the total NK response and its frequency and to test the significance of associations between VL and the percent contribution of the trifunctional NK response to the total NK cell response. The percentage of the parental 221 response for trifunctional 3DL1+ NK cells induced by each 221 HLA transfectant was calculated with the following formula: (frequency of Z27hi [i.e., 3DL1+] cells in a functional NK subset after incubation with a 221 transfectant/frequency of Z27hi cells in that NK functional subset after stimulation with the parental 221 cell line) × 100. A paired t test was used to assess the significance of the level of inhibition of the Z27hi trifunctional response between 221-HLA transfectants. A P value of <0.05 was considered significant.
RESULTS
NK cells from 3DL1 hmz SP who carry at least 1 HLA-Bw4 allele have higher trifunctional potential than those from Bw6 hmz.
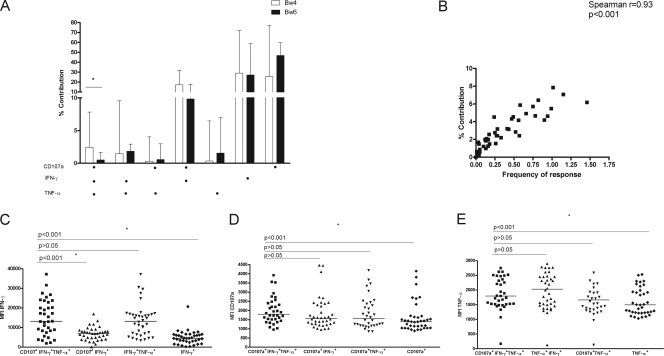
To investigate whether NK cells from HIV-infected SP grouped according to whether their 3DL1 and HLA-B alleles were NK receptor-HLA ligand pairs differ in their NK cell functional potential, we measured the expression of CD107a and secretion of IFN-γ and TNF-α from K562-stimulated NK cells from 47 HIV-infected 3DL1 hmz SP by using eight-color multiparametric flow cytometry. By measuring these three functions concurrently, we were able to assess the frequency of seven possible NK cell functional profiles that were positive responses to K562 stimulation and their percent contribution to the total K562 response. Figure S1A in the supplemental material shows the gating strategy that was used to obtain the frequency of each NK cell functional subset from which the percent contribution of each subset was calculated. Figure S1B in the supplemental material also shows examples of flow cytometry plots for NK cells from a representative *h/*y+B*57 SP stained with the isotype control for the Z27 monoclonal antibody that was used to set the gate for KIR3DL1+ NK cells (top row), an unstimulated control was used to set the gates for IFN-γ+, TNF-α+, and CD107a+ NK cells used to correct for background (middle row) and positive staining for IFN-γ, TNF-α, CD107a, and Z27 in a K562-stimulated test sample (bottom row). Figure S2 in the supplemental material shows flow cytometry plots of the frequency of trifunctional NK cells from representative *h/*y+B*57 and Bw6 individuals.
Figure 1A shows the percent contribution of each NK cell functional profile in PBMC from 3DL1 hmz expressing HLA-Bw4, the 3DL1 ligand (n = 40), compared with that from subjects who were Bw6 hmz (n = 7). Of the seven NK cell functional profiles, only the percent contribution of trifunctional NK cells to the total K562 response was significantly higher in the Bw4-positive SP compared to Bw6 hmz SP (medians of 2.42% [range, 0.0 to 7.8%] and 0.52% [range, 0.0 to 2.2%] for Bw4-positive and Bw6 hmz, respectively; corrected P = 0.032, Mann-Whitney U test). When the data were analyzed using the frequency of each NK functional subset, only the frequency of trifunctional cells was significantly higher in the Bw4-positive versus Bw6 hmz SP. Since in all the analyses performed, the frequency of each NK cell functional subset was correlated with its percent contribution to the total response (P < 0.0001; Spearman's r = 0.93) (Fig. 1B), we reported subsequent results as the percent contribution of a functional subset to the total NK cell response.
Fig. 1.
Stimulated NK cells from KIR3DL1 (3DL1) homozygous HIV-infected slow progressors (SP) have higher trifunctional activity when from carriers of an HLA-Bw4 allele than from Bw6 homozygotes (hmz). (A) Comparisons of percent contributions of seven functional profiles to the total NK response by NK cells from 3DL1 hmz SP stimulated with K562. Results for 40 SP carrying at least one Bw4 allele were compared to those for 7 Bw6 hmz. Dots below the x axis refer to the presence of a measured functional marker (CD107a, IFN-γ, and TNF-α). Bar heights refer to the median percent contribution of that functional subset to the total NK response for the group. Error bars refer to the upper range for the group. Significant between-group differences are shown with an asterisk over the line linking the two bars representing the groups being compared. (B) Correlation of results reported as the percent contribution of the trifunctional (CD107a+ IFN-γ+ TNF-α+) NK subset to the total NK response with results reported as the frequency of this functional NK subset for the 47 SP subjects in panel A. A Spearman's correlation test was used to test the significance of these two ways of reporting results. (C) Comparisons of the mean fluorescence intensities (MFI) of IFN-γ secretion are plotted on the y axis for trifunctional (CD107a+ IFN-γ+ TNF-α+), bifunctional (CD107a+ IFN-γ+ and IFN-γ+ TNF-α+), and monofunctional IFN-γ+ NK cells. Panel D shows the MFI of TNF-α secretion plotted on the y axis for trifunctional (CD107a+ IFN-γ+ TNF-α+), bifunctional (CD107a+ TNF-α+ and IFN-γ+ TNF-α+), and monofunctional TNF-α+ NK cells. Panel E shows the MFI of CD107a expression plotted on the y axis for trifunctional (CD107a+ IFN-γ+ TNF-α+), bifunctional (CD107a+ TNF-α+ and CD107a+ IFN-γ+), and monofunctional CD107a+ NK cells. For panels C, D, and E, the line through the scatter plots represents the median. Mann-Whitney U tests were used to test the significance of between-group comparisons. P values for these are shown above the lines linking two bars representing functional subsets.
We next questioned whether trifunctional NK cells were more potent than monofunctional cells for each function tested. We compared the MFI of IFN-γ and TNF-α secretion and CD107a expression in tri-, bi-, and monofunctional NK cells. As seen in Fig. 1C, the MFI of IFN-γ secretion from trifunctional NK cells was significantly higher than that from bifunctional CD107a+ IFN-γ+ NK cells and monofunctional NK cells secreting IFN-γ only (P < 0.001 for both comparisons, Mann-Whitney U test). There was no significant difference in the intensities of IFN-γ secretion between the trifunctional NK subset and the one that secreted IFN-γ and TNF-α (Fig. 1C). Figure 1D and E show the MFI of CD107 and TNF-α expression, respectively, in all of the NK subsets exhibiting these functions. Trifunctional NK cells expressed more CD107a and TNF-α than the monofunctional NK subsets with these activities (P < 0.001, Mann-Whitney U test). The amounts of CD107a and TNF-α secreted by trifunctional cells did not differ significantly from that secreted by the bifunctional subsets with these activities. Collectively these results indicate that on a per cell basis, trifunctional NK cells exhibit multiple functions, each with a higher intensity than monofunctional NK cells.
Receptor ligand requirements for increased NK cell trifunctional potential in *h/*y+B*57 SP.
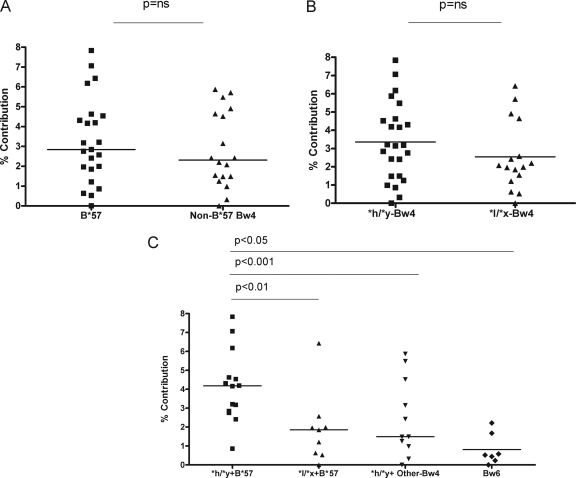
The 3DL1/Bw4 combination with the most potent influence on time to AIDS and VL set point is the *h/*y+B*57 genotype (36). Our previous work with HIV-uninfected subjects showed that NK cells from individuals with the *h/*y+B*57 genotype had higher NK trifunctional potential than those from carriers of the *h/*y+Bw4*80I, *h/*y+Bw4, or Bw6 hmz genotypes (9). The low frequency of B*57+ subjects from which these HIV-seronegative study subjects was drawn (28) and the even lower frequency of carriers of 3DL1*h/*y+B*57 or 3DL1*l/*x+B*57 precluded our being able to formally determine whether elevated trifunctional potential of NK cells from uninfected *h/*y+B*57 carriers required the presence of both the NK receptor and HLA ligand (9). B*57 is a protective allele in the context of HIV infection and is found at a higher frequency in HIV-infected SP than in the uninfected population. We were therefore able to assemble a sufficient number of SP expressing *h/*y+B*57, *h/*y with Bw4 alleles other than B*57 and *l/*x+B*57 carriers to address this point.
To assess the requirement for both carriage of a 3DL1*h/*y NK receptor genotype and HLA-B*57 ligand in elevated NK trifunctional potential, we compared the percent contributions of trifunctional NK cells from 23 3DL1 hmz who were B*57 positive to that of 18 3DL1 hmz who carried Bw4 alleles other than B*57 and found no significant between-group differences (P > 0.05, Mann-Whitney U test) (Fig. 2A). We next divided the same SP subjects into two groups based on their 3DL1 allotype. No significant between-group differences were observed in the percent contributions of trifunctional NK cells to the total NK response between 24 subjects who were 3DL1*h/*y and 17 subjects who were 3DL1*l/*x (P > 0.05, Mann-Whitney U test) (Fig. 2B).
Fig. 2.
Higher percent contribution of trifunctional NK cells to the total NK cell response in *h/*y+B*57 than Bw6 homozygotes (hmz) SP requires coexpression of the *h/*y 3DL1 receptor genotype and the HLA-B*57 ligand. Scatter plots show the percent contributions of trifunctional NK cells to the total K562-stimulated NK response for HLA-B*57 (B*57)-positive SP versus those with Bw4 alleles other than HLA-B*57 (Non-B*57-Bw4) (A), for carriers of HLA-Bw4 and 3DL1*h/*y genotypes (*h/*y+Bw4) versus 3DL1*l/*x genotypes (*l/*x+Bw4) (B), and for carriers of a 3DL1*h/*y genotype with HLA-B*57 (*h/*y+B*57), a 3DL1*l/*x genotype with HLA-B*57 (*l/*x+B*57), a 3DL1*h/*y genotype with an HLA-Bw4 allele other than HLA-B*57 (*h/*y-Other Bw4), and Bw6 hmz (C). The line through each scatter plot is the median for the group. Mann-Whitney U tests were used to test the significance of between-group differences. P values for comparison are shown over the line linking two groups.
The percent contribution of trifunctional NK cells from 14 *h/*y+B*57 carriers was higher than that for 9 *l/*x+B*57 carriers (median, 4.1% [range, 0.8 to 7.8%] versus 1.8% [range, 0.0 to 6.4%]; P < 0.01, Mann-Whitney U test) (Fig. 2C). This value was also higher than that seen in 12 individuals who carried the 3DL1*h/*y genotype with Bw4 alleles other than B*57 (*h/*y-Other Bw4) (median, 1.49% [range, 0.0 to 5.8%]), or 7 Bw6 hmz (median, 0.52% [range, 0.0 to 1.68%]; P = 0.05 and P < 0.001, respectively, Mann-Whitney U test) (Fig. 2C). Therefore, the enhanced NK trifunctional potential in *h/*y+B*57 individuals depends on carriage of both 3DL1*h/*y and B*57 and supports the interpretation that the expression of neither the NK receptor nor the HLA ligand alone is sufficient for the elevated NK trifunctional potential observed in carriers of the *h/*y+B*57 genotype.
The differences in trifunctional potential among these study groups were not due to differences in the percentage or absolute number of total NK cells or NK subsets such as CD56dim and CD56bright cells between SP grouped by 3DL1/HLA-B (see Fig. S3A to C in the supplemental material), nor did the percentages of 3DL1+ NK cells differ between groups (not shown). The only exception to this is that *h/*y+B*57 SP had significantly lower levels of the anergic CD56− CD16+ NK subset than SP with other 3DL1/HLA-B genotypes (see Fig. S3D in the supplemental material). In order to control for the possibility that increased trifunctional potential in NK cells from *h/*y+B*57 carriers was due to lower percentages of anergic CD56− CD16+ cells, we performed a subanalysis excluding the CD56− CD16+ subpopulation from the comparison of the groups shown in Fig. 2C. Removal of CD56− CD16+ cells from the analyses did not affect the conclusion that a significantly higher percent contribution of trifunctional NK cells to the total NK response was observed when NK cells were from carriers of *h/*y+B*57 in comparison to *l/*x+B*57, *h/*y+Bw4 alleles other than B*57, and Bw6 hmz genotypes (data not shown).
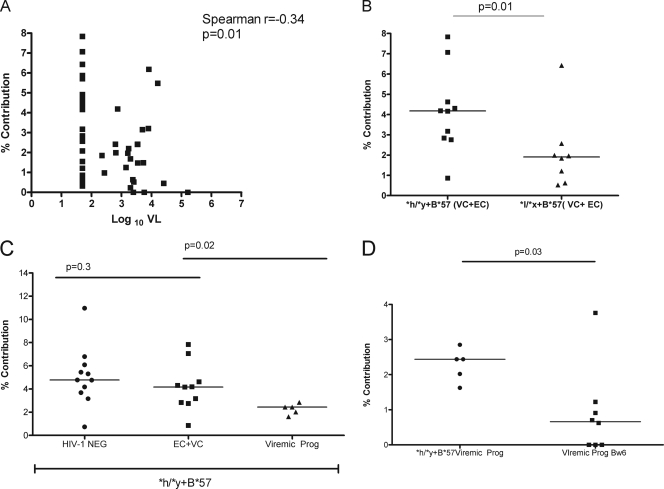
Influence of HIV VL on NK trifunctional potential.
3DL1 hmz SP stratified according to their 3DL1/HLA-B genotype exhibited subject-to-subject variation in their trifunctional potential (Fig. 2C). It was reported previously that HIV-1 infection dysregulates NK subset distribution and function (2, 5, 11, 37, 38). Our definition of SP in the study population included subjects with detectable VL. We therefore questioned whether there was evidence for any influence of VL on NK trifunctional potential. We observed a weak but significant negative correlation between the percent contribution of trifunctional NK cell responses to the total NK response and log10 VL (r = −0.34 and P = 0.01, Spearman's correlation test) (Fig. 3A). To address the possibility that EC with undetectable viremia were driving this negative correlation, we reanalyzed the association, excluding the EC subset. The significance of the correlation between log10 VL and NK cell trifunctional potential was lost (r = −0.03 and P = 0.88, Spearman's correlation test) (data not shown). No significant correlations were seen between the percent contribution of any of the bi- or monofunctional responses to the total K562-stimulated response and log10 VL (data not shown). In order to address the possibility that between-group differences in trifunctional potential could be accounted for by differences in VL, we compared VL in the four SP groups shown in Fig. 2C. No significant between-group differences in VL were observed (data not shown). To further address the concern that the differences in NK cell trifunctional potential in SP groups stratified according to their 3DL1/HLA-B genotypes could be attributed to differences in VL, we compared the trifunctional potential of NK cells from individuals with a VL below 3,000 copies/ml plasma (EC and VC) that were *h/*y+B*57 versus that in those that were *l/*x+B*57. We reasoned that the low levels of VL in these subjects would minimize the effect of VL on NK dysregulation. Although no significant differences were detected in the VL between EC and VC with *h/*y+B*57 and *l/*x+B*57 genotypes, carriers of *h/*y+B*57 had a significantly higher level of trifunctional NK cells than *l/*x+B*57 carriers (median, 4.17 [range, 0.86 to 7.83] and 1.90 [range, 0.52 to 6.43], respectively; P = 0.01, Mann-Whitney U test) (Fig. 3B).
Fig. 3.
Influence of HIV viral load (VL) and KIR/HLA genotype on NK functional potential. (A) Correlation between the percent contribution of trifunctional NK cells to the total K562-stimulated NK response and log10 VL. A Spearman's correlation test was used to assess the significance of the association between these two parameters. (B) Scatter plots show the distribution of the percent contribution of trifunctional NK cells to the total K562-stimulated NK response in HIV-infected individuals meeting the criteria for classification as either elite controllers (EC; <50 HIV copies/ml plasma) or viral controllers (VC; <3,000 HIV copies/ml plasma) that carry a 3DL1*h/*y genotype with HLA-B*57 (*h/*y-B*57) versus a 3DL1*l/*x genotype with HLA-B*57 (*l/*x-B*57). (C) Scatter plots show the distribution of the percent contribution of trifunctional NK cells to the total K562-stimulated NK response in HIV-1-negative individuals, HIV-1-infected EC and VC, and untreated viremic progressors matched for the *h/*y+B*57 genotype. (D) Scatter plots are the same as in panel C for untreated viremic progressors carrying the *h/*y+B*57 genotype versus the Bw6 hmz genotype. For panels B to D, the line through each scatter plot is the median for the group. Mann-Whitney U tests were used to test the significance of between-group differences. P values for comparison are shown over the line linking two groups.
We next compared NK cell trifunctional potentials in three groups of individuals matched for the *h/*y+B*57 combined genotype, 11 HIV-uninfected subjects, 10 HIV-infected individuals with a VL of <3,000 copies/ml plasma (EC and VC), and 5 viremic HIV-infected progressors. Whereas no significant differences were found between HIV-negative individuals and EC and VC with this genotype combination (median, 4.8 [range, 0.7 to 11] versus 4.17 [range, 0.86 to 7.83]; P = 0.3, Mann-Whitney U test), the EC and VC had significantly higher levels of trifunctional NK cells versus viremic progressors (median, 2.43 [range, 1.62 to 2.85]; P = 0.02, Mann-Whitney U test) (Fig. 3C). Furthermore, *h/*y+B*57 viremic progressors had significantly higher levels of trifunctional NK cells than eight Bw6 viremic progressors (median, 0.66 [range, 0.00 to 3.87]; P = 0.03, Mann-Whitney U test), even though these groups did not differ significantly from each other for VL (P > 0.05, Mann-Whitney U test) (Fig. 3D). Together, these results suggest that even though the negative impact of VL on NK function may contribute to some of the variability in NK polyfunctional potential, 3DL1/HLA-B genotype combinations are also determinants of NK functional potential.
3DL1+ NK responses from *h/*y+B*57 SP are inhibited by 221 cells bearing B*5701 and B*2702 alleles.
In order to determine whether the NK activation seen following stimulation with HLA-devoid cells was related, in part, to the loss of suppressive signals mediated 3DL1 interacting with its ligand, we stimulated NK cells from individuals expressing the *h/*y+B*57 genotype with the HLA-devoid 221 cell line and a panel of 221 transfectants expressing single HLA alleles (namely; the Bw4*80I alleles B*57:01 and B*27:02 and B*35:02, a Bw6 allele). For these experiments, we used the Z27 MAb to distinguish the 3DL1+ from 3DL1− NK cells.
Figure 4A shows flow cytometry plots of the frequency of 3DL1+ and 3DL1− NK cells secreting IFN-γ following stimulation of PBMC from a representative *h/*y+B*57 carrier with medium alone, the parental 221 cell line, and 221 HLA transfectants. Stimulation with 221 induced 16% of 3DL1+ NK cells to secrete IFN-γ, whereas 221-B*35:02 induced 11.4% of these cells to secrete this cytokine. Stimulation with 221-B*57:01 and 221-B*27:02 suppressed IFN-γ secretion to 1.46% and 5.75% of that of the 3DL1+ NK cells, respectively. Within the 3DL1− compartment, the extent of suppression of IFN-γ secretion by 221-B*57:01 and 221-B*27:02 compared to that by 221 and 221-B*35:02 target cells was much less.
Fig. 4.
Cell line 721.221 (221) transfectants expressing HLA-B*57:01 and B*27:02 but not B*35:02 suppress 3DL1+NK cell function. (A) Flow cytometry plots of peripheral blood mononuclear cells (PBMC) from a representative individual carrying the 3DL1*h/*y genotype with HLA-B*57 (*h/*y+B*57) following stimulation with medium alone, the parental 221 cell line, and 221 cells transfected with single HLA alleles. The 221 stimulus is indicated over the plots with the parental cell line designated 221, 221-B*57:01 as B*57, 221-B*27:02 as B*27, and 221-B*35:02 as B*35. NK cells staining positive for the Z27 monoclonal antibody (3DL1+) cells were gated in the upper panels, and Z27− (3DL1−) NK cells were gated in the lower panels. The number in each plot shows the frequency of the IFN-γ+ 3DL1+ (upper) or 3DL1− (lower) response in the boxed area following stimulation. Panels B to D show pooled data from 10 *h/*y+B*57 SP. Panels B and C show the results for the percent contribution of total IFN-γ secretion within the 3DL1+ (B) and 3DL1− (C) compartments, respectively. Mann-Whitney U tests assessed the significance of differences in the percent contributions of IFN-γ to the 3DL1+ response and 3DL1− response upon stimulation with 221-B*57:01, 221-B*27:02, and 221-B*35:02 transfectants versus the 221 parental cell line. Panel D shows results from 10 *h/*y+B*57 subjects for the trifunctional 3DL1+ NK response following culture with the 221 transfectant panel expressed as a percentage of the response to the 221 parental cell line. Paired t tests were used to test the significance of the frequencies of trifunctional 3DL1+ NK cells upon stimulation with 221-B*57:01 and 221-B*27:02 transfectants versus the 221-B*35:01 transfectant. For panels B to D, bar and error bar lengths represent the mean and standard error for the group. *, P < 0.05; **, P < 0.01.
Figure 4B and C show pooled results for the percent contribution of IFN-γ+-secreting 3DL1+ and 3DL1− NK functional subsets to the total 3DL1+ and 3DL1− NK cell response, respectively, from 10 *h/*y+B*57 carriers stimulated with the 221 panel. The percent contributions of IFN-γ+ 3DL1+ NK cells to the 3DL1+ NK cell response were significantly lower following stimulation with 221-B*57:01 and 221-B*27:02 than that with the parental 221 cell line (P < 0.01 for both comparisons, Mann Whitney U test). The percent contributions of IFN-γ+-secreting 3DL1+ cells to the 3DL1+ response did not differ between the 221-B*35:02 and the 221 parental line. None of the 221 transfectants suppressed IFN-γ secretion within the 3DL1− NK compartment compared to the parental 221 cell line. These results confirm that the loss of the interaction between 3DL1 receptors and their ligands is important to the enhanced function of NK cells from *h/*y+B*57 carriers following stimulation with HLA-devoid cells. The presence of either B*57:01 or B*27:02 ligands on 221 transfectants that can interact with 3DL1 receptors is sufficient to suppress NK function in the NK subset expressing this receptor; in contrast, 221-B*35:02 transfectants do not suppress NK cell function of 3DL1+ cells. Neither the 221 parental line nor any of the 221 transfectants suppressed NK function in the 3DL1− compartment.
We next examined the effect of the three transfectants on the 3DL1+ trifunctional NK cell response. In Fig. 4D, the effect of each HLA tranfectant on the 3DL1+ trifunctional subset is expressed as a percentage of the response to the parental 221 cell line, which is set at 100%. The 221-B*57:01 and 221-B*27:02 transfectants suppressed the trifunctional activity of 3DL1+ NK cells, whereas the 221-B*3502 tranfectant did not. Together, these results indicate that interactions between cells expressing single HLA Bw4*80I alleles and 3DL1+ NK cells suppress NK activity.
DISCUSSION
Of the KIR/HLA combinations associated with slow time to AIDS and VL control, *h/*y+B*57 is the most potent (36). We report here that in 3DL1 hmz SP with at least 1 copy of a Bw4 allele, trifunctional NK cells contribute significantly more to the total K562-stimulated response than those from Bw6 hmz. *h/*y+B*57 carriers have the highest NK trifunctional potential compared with carriers of other 3DL1 hmz receptor genotypes and/or Bw4 alleles. Trifunctional NK cells have a higher functional potency than corresponding monofunctional cells. Although VL negatively impacts NK trifunctional potential, VL alone does not account for variations in NK functional potential. We were able to confirm that HLA-devoid cell line stimulation of NK trifunctional potential in *h/*y+B*57 carriers arises at least in part from the abrogation of inhibitory signals mediated by 3DL1 by showing that stimulation with 221 HLA transfectants expressing Bw4*80I alleles, but not Bw6 alleles, suppressed the function of 3DL1+ NK cells from these individuals.
The more potent effector functions mediated by polyfunctional NK cells compared to corresponding monofunctional subsets is reminiscent of that seen in polyfunctional CD8+ T cells (6, 16). Polyfunctional CD8+ T cells are commonly observed in the setting of effectively controlled viral infections such as cytomegalovirus, EBV, vaccinia virus, and influenza virus infections (23, 56). The preferential maintenance of multifunctional HIV-specific CD8+ T cells in HIV-infected long-term nonprogressors (LTNP) compared to progressors may be due to polyfunctional CD8+ T cells having superior anti-HIV activity or the effect of low viremia limiting the functional exhaustion of these cells (6, 34). We report here a similar phenomenon for trifunctional versus monofunctional NK cells. We did not detect significant differences in the MFI of TNF-α secretion and CD107a expression between the bifunctional and trifunctional NK cell subsets. The bifunctional CD107a+ IFN-γ+ NK subset, however, secreted smaller amounts of IFN-γ than the trifunctional NK cells. Whether trifunctional NK cells are endowed with a superior capacity to suppress viral replication in infected targets than monofunctional NK cells warrants further investigation. In the work reported here, we have not directly tested the antiviral activity of NK cells from subjects with defined 3DL1/HLA-B genotypes since we did not use HIV-infected cells as targets. Although the biological relevance of polyfunctional antigen-specific CD8+ T cells in HIV infection is not yet clear, they do serve as an indicator of an effective response to HIV. Our data illustrate that NK cells with polyfunctional potential may also be of immunological relevance in this context.
The percent contribution of trifunctional responses to the total NK cell response varied within study populations stratified by 3DL1/HLA-B. It is possible that some of this variation is due to VL since VL was negatively, albeit weakly, correlated with trifunctional potential. The finding that *h/*y+B*57 HIV viremic progressors have lower levels of NK trifunctional potential than *h/*y+B*57-matched HIV-uninfected and HIV-infected EC and VC would be consistent with the disruption of NK cell function in HIV-infected individuals with uncontrolled viremia, as has been reported by others (1, 2). However, VL is unlikely to be the only determinant of NK functional potential since there were no significant differences in the VL of SP grouped by the 3DL1/HLA-B genotypes. Furthermore, in a subset of EC and VC where the effect of VL would be minimized, we found significantly higher NK trifunctional potential when cells were from *h/*y+B*57 subjects than when they were from *l/*x+B*57 subjects. Our data do not dispute that HIV infection affects NK cell function but provide evidence that 3DL1/HLA-B genotype is also a determinant of NK functional potential. We were unable to examine and compare the NK trifunctional potentials in successfully treated individuals matched for the *h/*y+B*57 combined genotype, because we identified only two such individuals.
Functional heterogeneity of NK cell responses within *h/*y+B*57 carriers and other groups classified by their 3DL1/HLA-B genotype may arise through the contribution of KIR/HLA receptor-ligand combinations other than 3DL1/Bw4 to the NK functional repertoire (40, 55). The number of inhibitory KIR to self HLA alleles a person carries, the HLA-B alleles carried (Bw6, Bw4, or Bw4*80I), the frequency of KIR3DL1+ NK cells, and level of expression of KIR3DL1 on NK cells may all contribute to variability in NK functional potential within groups classified by 3DL1/HLA-B genotype (53, 55). HLA and KIR region typing to identify the presence of other KIR-HLA combinations that have an impact on NK education would be a first step to addressing the contribution of other KIR/HLA combinations to NK functional potential. Expansion of the antibody panel to detect NK subsets expressing single or defined combinations of other NK receptors, use of blocking antibodies for NK receptors, and a broader array of 221 transfectants will further aid in dissecting the contribution of other KIR-HLA combinations to stimulated NK function.
Interactions between HIV and NK cells lead to the pathogenic redistribution of NK cells in the peripheral blood of HIV-infected subjects (2, 37, 38). The expansion of a dysfunctional anergic CD56− CD16+ NK cell population has been associated with high VL during the chronic phase of HIV infection. In HIV-infected persons with low VL, this dysfunctional subset is smaller than that seen in progressors. Suppression of VL by effective antiretroviral therapy also leads to a reduction in the size of this subset (2, 11). The percentages or absolute numbers of total NK cells and CD56dim, CD56bright, and KIR3DL1+ NK cell subpopulations were similar in the SP groups categorized by 3DL1/HLA-B. However, we did find a significantly lower frequency of the anergic CD56− CD16+ NK cells in the *h/*y+B*57 carriers than that in SP with any other 3DL1/HLA-B genotype. Excluding this anergic subset in a subanalysis confirmed that increased NK trifunctional potential was not due to the decreased frequency of CD56− CD16+ among *h/*y+B*57 SP.
As has been reported before for uninfected individuals, stimulated NK cells from HIV-infected *h/*y+B*57-positive SP have higher levels of trifunctional NK cells than carriers of other 3DL1/HLA-B combinations such as *l/*x+B*57, *h/*y+other Bw4 alleles, or Bw6 hmz (9). In the case of the *l/*x+B*57 combination, the strength of the interaction between KIR receptors and B*57 may be weaker than that between 3DL1 receptors and the HLA ligand in *h/*y+B*57 subjects. Although this may depend on individual 3DL1 receptors and HLA ligands, in general, the receptors in the 3DL1*l/*x genotype are expressed on the NK cell surface at lower levels, and several members of the *l 3DL1 group have been shown to be weaker inhibitory alleles than common *h 3DL1 alleles (12, 52). In the case of Bw6 hmz, none of the Bw6 alleles are ligands for 3DL1 receptors. Therefore, NK cells from Bw6 hmz are not expected to receive educating signals through 3DL1. The difference in functional potential between NK cells from carriers of *h/*y+B*57 versus *h/*y+other Bw4 alleles may reflect differences in the impact of B*57 alleles versus other Bw4 alleles in providing educating signals to NK cells during development. Transgenic mice expressing single MHC class I alleles have shown that MHC class I alleles can differ in their impacts on NK cell education (24, 25, 45). This process is important for the development of self-tolerant NK cells and for endowing NK cells with the capacity to mediate cytokine secretion and cytolysis upon encountering virally infected cells or tumor cells with aberrant MHC class I expression or with ligands for activating NK receptors (27). The strength of the inhibitory input during education is an important factor in determining the functional responsiveness of individual NK cells (10, 25). One interpretation of the finding that carriers of the *h/*y+other Bw4 combinations had NK cells with a lower functional potential than those from *h/*y+B*57 carriers is that B*57 differs from most other Bw4 alleles in terms of the strength with which it interacts with 3DL1 to educate NK cells. Although there is experimental evidence that Bw4*80I alleles interact with 3DL1 more strongly than Bw4*80T alleles, there has been no direct examination of the strength with which B*57 binds any 3DL1 allele compared to other Bw4 alleles (13, 18, 22).
221 cells expressing single HLA alleles allowed us to confirm that NK activation following stimulation with HLA-devoid cells was in part due to disruption of the inhibitory signal mediated by 3DL1 binding to its ligand. Stimulation of NK cells from *h/*y+B*57 carriers with both 221-B*57:01 and 221-B*27:02 Bw4*80I transfectants, but not the 221-B*35:02 Bw6 transfectant, suppressed IFN-γ secretion within the 3DL1+ but not the 3DL1− NK cell compartment. The 221 and HLA transfectant panel allowed us to confirm this for the 3DL1+ trifunctional NK cell response as well.
Host factors associated with the control of HIV infection may help identify important targets for vaccine design. In this report, we measured NK functions that can mediate antiviral activities (31, 32, 39, 48, 51). Although we have not directly tested the antiviral activity of NK cells from subjects carrying *h/*y+B*57 since we did not use HIV-infected cells to stimulate NK responses, the results presented here may be relevant to HIV control. NK responses induced upon encountering HIV-infected cells with reduced HLA-A and -B expression may be one of the mechanisms underlying reduced risk of HIV infection and slower disease course in carriers of the *h/*y+B*57 genotype who become infected (14, 15). The results presented would be consistent with the possibility that SP who express the protective B*57 allele may be able to control HIV not only through CD8+ T-cell recognition of HIV peptides restricted by the B*57 alleles but also through interactions between B*57 and 3DL1 receptors that educate NK cells for potent antiviral activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank the study participants of the Canadian Cohort of HIV Infected Slow Progressors and the National Institute of Allergy and Infectious Diseases Cohort. We are grateful to Mario Legault and Stephanie Matte for cohort coordination. We thank Matthew Parsons for critical review of the manuscript. We also acknowledge the expert technical support of Marie-Pierre Boisvert, Tsoarello Mabanga, and Saied Sharafi.
This work was supported by grant HOP-86862 from the Canadian Institutes for Health Research, Canadian Association for AIDS Research grant 022-110, and a grant from the Fonds de Recherche en Santé du Québec (FRSQ) AIDS and Infectious Disease Network. S.B. was supported by a Ph.D. scholarship from the FRSQ, and J.-P.R. is a scientific scholar receiving support from FRSQ.
The following are contributing members of the Canadian Cohort of HIV Infected Slow Progressors: Bernard Lessard, Clinique Quartier Latin, Montréal, Québec, Canada; Danielle Legault, Pierre-Jean Maziade, Danièle Longpré, Sylvie Vézina, and Benoit Trottier, Clinique L'Actuel, Montréal, Québec, Canada; Danielle Rouleau, Centre de Recherche du Centre Hospitalier de l'Université de Montréal, Montréal, Québec, Canada; Howard Turner and Julian Falutz, Faculty of Medicine, Department of Medicine, McGill University, Montreal, Quebec, Canada; Martin Potter, Research Institute of the McGill University Health Centre, Montréal, Québec, Canada; and Marina B. Klein, McGill University Health Centre, Division of Infectious Diseases and Immunodeficiency Service, Montréal, Québec, Canada.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Alter G., et al. 2003. Longitudinal assessment of changes in HIV-specific effector activity in HIV-infected patients starting highly active antiretroviral therapy in primary infection. J. Immunol. 171:477–488 [DOI] [PubMed] [Google Scholar]
- 2. Alter G., et al. 2005. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood 106:3366–3369 [DOI] [PubMed] [Google Scholar]
- 3. Altfeld M., et al. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bancroft G. J. 1993. The role of natural killer cells in innate resistance to infection. Curr. Opin. Immunol. 5:503–510 [DOI] [PubMed] [Google Scholar]
- 5. Barker E., Martinson J., Brooks C., Landay A., Deeks S. 2007. Dysfunctional natural killer cells, in vivo, are governed by HIV viremia regardless of whether the infected individual is on antiretroviral therapy. AIDS 21:2363–2365 [DOI] [PubMed] [Google Scholar]
- 6. Betts M. R., et al. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boulassel M. R., et al. 2003. Changes in immunological and virological parameters in HIV-1 infected subjects following leukapheresis. J. Clin. Apher. 18:55–60 [DOI] [PubMed] [Google Scholar]
- 8. Boulet S., et al. 2008. Increased proportion of KIR3DS1 homozygotes in HIV-exposed uninfected individuals. AIDS 22:595–599 [DOI] [PubMed] [Google Scholar]
- 9. Boulet S., et al. 2010. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J. Immunol. 184:2057–2064 [DOI] [PubMed] [Google Scholar]
- 10. Brodin P., Lakshmikanth T., Johansson S., Karre K., Hoglund P. 2009. The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells. Blood 113:2434–2441 [DOI] [PubMed] [Google Scholar]
- 11. Brunetta E., et al. 2009. The decreased expression of Siglec-7 represents an early marker of dysfunctional natural killer-cell subsets associated with high levels of HIV-1 viremia. Blood 114:3822–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carr W. H., Pando M. J., Parham P. 2005. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 175:5222–5229 [DOI] [PubMed] [Google Scholar]
- 13. Cella M., Longo A., Ferrara G. B., Strominger J. L., Colonna M. 1994. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J. Exp. Med. 180:1235–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cohen G. B., et al. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661–671 [DOI] [PubMed] [Google Scholar]
- 15. Collins K. L., Chen B. K., Kalams S. A., Walker B. D., Baltimore D. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397–401 [DOI] [PubMed] [Google Scholar]
- 16. Darrah P. A., et al. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13:843–850 [DOI] [PubMed] [Google Scholar]
- 17. Degli-Esposti M. A., Smyth M. J. 2005. Close encounters of different kinds: dendritic cells and NK cells take centre stage. Nat. Rev. Immunol. 5:112–124 [DOI] [PubMed] [Google Scholar]
- 18. Draghi M., et al. 2005. Single-cell analysis of the human NK cell response to missing self and its inhibition by HLA class I. Blood 105:2028–2035 [DOI] [PubMed] [Google Scholar]
- 19. Fellay J., et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gardiner C. M., et al. 2001. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J. Immunol. 166:2992–3001 [DOI] [PubMed] [Google Scholar]
- 21. Goulder P. J., et al. 1996. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retroviruses 12:1691–1698 [DOI] [PubMed] [Google Scholar]
- 22. Gumperz J. E., Litwin V., Phillips J. H., Lanier L. L., Parham P. 1995. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J. Exp. Med. 181:1133–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harari A., et al. 2008. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J. Exp. Med. 205:63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoglund P., et al. 1988. Natural resistance against lymphoma grafts conveyed by H-2Dd transgene to C57BL mice. J. Exp. Med. 168:1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johansson S., et al. 2005. Natural killer cell education in mice with single or multiple major histocompatibility complex class I molecules. J. Exp. Med. 201:1145–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaslow R. A., et al. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405–411 [DOI] [PubMed] [Google Scholar]
- 27. Kim S., et al. 2008. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc. Natl. Acad. Sci. U. S. A. 105:3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lalonde R. G., et al. 2010. Successful implementation of a national HLA-B*5701 genetic testing service in Canada. Tissue Antigens 75:12–18 [DOI] [PubMed] [Google Scholar]
- 29. Lanier L. L. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274 [DOI] [PubMed] [Google Scholar]
- 30. Leslie A. J., et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289 [DOI] [PubMed] [Google Scholar]
- 31. Levy D. E., Garcia-Sastre A. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143–156 [DOI] [PubMed] [Google Scholar]
- 32. Lichterfeld M., et al. 2004. HIV-1-specific cytotoxicity is preferentially mediated by a subset of CD8(+) T cells producing both interferon-gamma and tumor necrosis factor-alpha. Blood 104:487–494 [DOI] [PubMed] [Google Scholar]
- 33. Litwin V., Gumperz J., Parham P., Phillips J. H., Lanier L. L. 1994. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J. Exp. Med. 180:537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Makedonas G., Betts M. R. 2011. Living in a house of cards: re-evaluating CD8+ T-cell immune correlates against HIV. Immunol. Rev. 239:109–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin M. P., et al. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429–434 [DOI] [PubMed] [Google Scholar]
- 36. Martin M. P., et al. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 39:733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mavilio D., et al. 2003. Natural killer cells in HIV-1 infection: dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc. Natl. Acad. Sci. U. S. A. 100:15011–15016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mavilio D., et al. 2005. Characterization of CD56−/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc. Natl. Acad. Sci. U. S. A. 102:2886–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mestan J., et al. 1986. Antiviral effects of recombinant tumour necrosis factor in vitro. Nature 323:816–819 [DOI] [PubMed] [Google Scholar]
- 40. Middleton D., Gonzelez F. 2010. The extensive polymorphism of KIR genes. Immunology 129:8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Migueles S. A., et al. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Migueles S. A., et al. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miura T., et al. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare Gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J. Virol. 83:2743–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Norman P. J., et al. 2007. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat. Genet. 39:1092–1099 [DOI] [PubMed] [Google Scholar]
- 45. Ohlen C., et al. 1989. Prevention of allogeneic bone marrow graft rejection by H-2 transgene in donor mice. Science 246:666–668 [DOI] [PubMed] [Google Scholar]
- 46. Pando M. J., Gardiner C. M., Gleimer M., McQueen K. L., Parham P. 2003. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J. Immunol. 171:6640–6649 [DOI] [PubMed] [Google Scholar]
- 47. Sanabria M. X., Vargas-Inchaustegui D. A., Xin L., Soong L. 2008. Role of natural killer cells in modulating dendritic cell responses to Leishmania amazonensis infection. Infect. Immun. 76:5100–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schroder K., Hertzog P. J., Ravasi T., Hume D. A. 2004. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75:163–189 [DOI] [PubMed] [Google Scholar]
- 49. Trinchieri G. 1989. Biology of natural killer cells. Adv. Immunol. 47:187–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wan A. M., Ennis P., Parham P., Holmes N. 1986. The primary structure of HLA-A32 suggests a region involved in formation of the Bw4/Bw6 epitopes. J. Immunol. 137:3671–3674 [PubMed] [Google Scholar]
- 51. Wong G. H., Goeddel D. V. 1986. Tumour necrosis factors alpha and beta inhibit virus replication and synergize with interferons. Nature 323:819–822 [DOI] [PubMed] [Google Scholar]
- 52. Yawata M., et al. 2006. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 203:633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yawata M., et al. 2008. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood 112:2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yokoyama W. M., Kim S. 2006. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol. Rev. 214:143–154 [DOI] [PubMed] [Google Scholar]
- 55. Yu J., et al. 2007. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J. Immunol. 179:5977–5989 [DOI] [PubMed] [Google Scholar]
- 56. Zimmerli S. C., et al. 2005. HIV-1-specific IFN-gamma/IL-2-secreting CD8 T cells support CD4-independent proliferation of HIV-1-specific CD8 T cells. Proc. Natl. Acad. Sci. U. S. A. 102:7239–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.