Abstract
Natural infection with simian retrovirus (SRV) has long been recognized in rhesus macaques (RMs) and may result in an AIDS-like disease. Importantly, SRV infections persist as a problem in recently imported macaques. Therefore, there is a clear need to control SRV spread in macaque colonies. We developed a recombinant vesicular stomatitis virus (VSV)-SRV vaccine consisting of replication-competent hybrid VSVs that express SRV gag and env in separate vectors. The goal of this study was to assess the immunogenicity and protective efficacy of the VSV-SRV serotype 2 vaccine prime-boost approach in RMs. The VSV-SRV vector (expressing either SRV gag or env) vaccines were intranasally administered in 4 RMs, followed by a boost 1 month after the first vaccination. Four RMs served as controls and received the VSV vector alone. Two months after the boost, all animals were intravenously challenged with SRV-2 and monitored for 90 days. After the SRV-2 challenge, all four controls became infected, and viral loads (VLs) ranged from 106 to 108 SRV RNA copies/ml of plasma. Two animals in the control group developed simian AIDS within 7 to 8 weeks postinfection and were euthanized. Anemia and weight loss were observed in the remaining controls. During acute infection, severe B-cell depletion and no significant changes in T-cell population were observed in the control group. Control RMs with greater preservation of B cells and lower VLs survived longer. SRV-2 was undetectable in vaccinated animals, which remained healthy, with no clinical or biological signs of infection and preservation of B cells. Our study showed that the VSV-SRV vaccine is a strong approach for preventing clinically relevant type D retrovirus infection and disease in RMs, with protection of 4/4 RMs from SRV infection and prevention of B-cell destruction. B-cell protection was the strongest correlate of the long-term survival of all vaccinated and control RMs.
INTRODUCTION
Simian retroviruses (SRV) cause an immunosuppressive disease in macaques (25, 26) and were a leading cause of fatal disease in macaque colonies in the United States (7). The problem persists as SRV infections have been found in recently imported macaques (42). The inadvertent use of SRV-infected nonhuman primates (NHPs) in biomedical research may severely affect animal studies (12). SRV testing and eradication are therefore an essential component of the definition of specific-pathogen-free (SPF) colonies. Additionally, control of SRV spread also improves safety for researchers and animal handlers by reducing the risk of transmission of a potentially pathogenic retrovirus (22).
SRV-1 is a pathogen specific to rhesus macaques (RMs), while SRV-2 may infect RMs as well as cynomolgus and pig-tailed macaques (PTMs) (21). Although a proportion of animals recover and control viral replication, others remain healthy carriers and can transmit virus by biting, through saliva (11). While effective immune control in some individuals may limit transmission of SRV-2 within a colony, it highlights the occurrence of latently infected animals from which reactivated virus may be transmitted under certain circumstances through various routes of transmission (21).
The fact that experimental infection of RMs with SRV-1 revealed that certain animals can overcome SRV infection with no clinical signs indicates the feasibility of developing successful vaccines (19). Indeed, the first vaccine for SRV-1 was formalin-inactivated vaccine grown in human cells that protected RMs against lethal challenge (26). SRV-1 challenge virus was grown in RM cells, thereby avoiding anticellular antibody (Ab) induction. However, another experimental vaccine that used SRV-1 gp70 and gp20 expressed in Saccharomyces cerevisiae failed to elicit neutralizing antibodies (18). Recombinant vaccinia virus expressing SRV-2 envelope glycoprotein protected PTMs against lethal challenge (2, 14). This protection correlated with the development of neutralizing antibodies. In another study, recombinant vaccinia virus expressing SRV-1 or SRV-3 (Mason-Pfizer monkey virus) gp70 and gp20 were shown to protect against homologous virus. Although a degree of cross-neutralization was observed among SRV-1- and SRV-3-vaccinated animals, they failed to cross neutralize the more distantly related SRV-2 (3). Collectively, these data show that a vector strategy that can be employed to immunize animals for SRV in primate facilities is feasible.
Vesicular stomatitis virus (VSV) is a nonsegmented, negative-strand RNA virus whose viral genome can be easily manipulated, allowing the insertion of foreign genes (20, 38). VSV vectors have been used for the development of vaccine against lentiviruses, including human immunodeficiency virus (HIV) (10, 16, 17). Furthermore, multiple strains of recombinant VSVs (rVSVs) have also been used in oncolytic viral therapy (6, 23, 39). More than 150 NHPs have been inoculated with rVSV vectors via intranasal (i.n.) and intramuscular routes, with no adverse effects (5, 36).
We developed a new vaccination strategy for SRV by use of VSV vectors. We constructed recombinant VSVs expressing the SRV-2 Gag and Env glycoproteins and employed a prime-boost approach utilizing two different recombinant VSVs expressing the Indiana and Chandipura G glycoproteins for priming and boosting immunizations, respectively. We report that all vaccinees from this study remained healthy, with undetectable VLs for up to 3 months postchallenge, while two out of four control animals progressed to AIDS, with an average time of about 7 weeks, and were euthanized. We also report that SRV challenge of RMs induced CD20+ B-cell depletion in control RMs but not in vaccinated macaques. These results suggest that VSV recombinant vaccines expressing the SRV-2 genes may efficiently prevent type D retrovirus-induced immunosuppressive disease in macaques.
MATERIALS AND METHODS
Cloning of SRV-2 gag and env genes.
gag and env genes were PCR amplified from the plasma of an SRV-2-positive animal and were cloned into a p3x-FLAG expression vector (Sigma, St. Louis, MO) as follows. NotI and BamHI sites were introduced at the 5′ and 3′ ends, respectively, using the 5′-GAG-FLAG-NotI and 3′-GAG-FLAG-BamHI primers (see Table S1 in the supplemental material). To construct the SRV-2 Env/VSV G cytoplasmic tail fusion, portions of the individual genes were PCR amplified and the fragments were later fused by using PCR overlap extension (Fig. 1). Briefly, a C terminus-truncated portion of the SRV-2 envelope gene was based on the 38-amino-acid (aa) cytoplasmic domain of SRV-3 (4) and also estimated using the SOSUI algorithm (13, 30). The SRV-2 Env region (bases 1 to 1638) was PCR amplified using 5′-ENV-FLAG-NotI and 3′-SOLENV-FLAG-BamHI while the region encoding the last 29 amino acids of the VSV G cytoplasmic tail (bases 1447 to 1533) (34, 35) was PCR amplified using 5′-G-CYT-ENV-JN and 3′-G-CYT-BamHI. In a final PCR, these two fragments were fused by a PCR overlap extension technique using the 5′-ENV-FLAG-NotI and 3′-XN2-NheI primers. The constructs encoding the truncated portion of ENV in-frame with the 3× FLAG protein was termed ENVTrunc (Fig. 1B), and the construct with the VSV-G cytoplasmic tail fusion was termed EnvGCYT (Fig. 1A).
Fig. 1.
Schematic depicting SRV-2 EnvGCYT and EnvTrunc constructs. The secreted portion of the envelope glycoprotein (nucleotides [nt] 1 to 1638) was fused in-frame to the VSV G cytoplasmic tail (nt 1447 to 1533) and FLAG epitope for the EnvGCYT construct (A) or to directly to the FLAG epitope alone for the EnvTrunc construct (B).
Construction and recovery of recombinant VSVs expressing the SRV-2 gag and env genes.
For the SRV-2 gag construct, the plasmid clone that efficiently expressed the gag gene was used as the template for PCR amplification of the gene, while at the same time introducing unique XhoI and NheI sites at the 5′ and 3′ ends of the gene fragment. The 5′-XN2-GAG-XhoI and 3′-XN2-NheI primers (see Table S1 in the supplemental material) were used for this process. This DNA fragment was cloned into the pVSV-XN2 Indiana and pVSV-XN2 Chandipura transfer vectors (kindly provided by John Rose). Similarly, EnvTrunc and EnvGCYT were PCR amplified using the 5′-XN2-ENV-SalI and 3′-XN2-NheI primers (Table S1) before being cloned into the pVSV-XN2 Indiana and pVSV-XN2 Chandipura transfer vectors. The recombinant VSVs were recovered as described previously (15). Briefly, BHK 21 cells were infected with recombinant vaccinia virus expressing T7 polymerase (vTF7-3) at a multiplicity of infection (MOI) of 10 for 1 h. Subsequently, the cells were cotransfected with pBS-N, pBS-P, pBS-L, and pVSV-XN2 containing either the Gag, the EnvTrunc, or the EnvGCYT gene, and recombinant virus was recovered through multiple rounds of filtration steps. Control viruses having no exogenous inserted genes were also generated using pBS-N, pBS-P, pBS-L, and pVSV-XN2 (empty vector). Viral isolates expressing large amounts of the SRV proteins were selected through multiple rounds of plaque purification. Viral titers were determined, and stocks were stored at −80°C for vaccination studies.
Detection of SRV-2 Gag, EnvTrunc, and EnvGCYT expression by rVSVs.
The VSV–SRV-2 Gag, VSV–SRV-2–ENVTrunc, and VSV–SRV-2–EnvGCYT recombinants were tested for expression by infecting BHK 21 cells. Gag constructs were probed in a Western immunoblot analysis using anti-FLAG antibodies (Sigma) and detected using horseradish peroxidase (HRP)-conjugated goat anti-mouse antibodies (Molecular Probes, Eugene, OR). The Env constructs were probed using pooled sera from five SRV-2-positive macaques as primary antibodies and HRP-conjugated goat anti-monkey (Fitzgerald industries, North Acton, MA) secondary antibodies.
Animals.
Eight Indian-origin RMs were used. All animals were negative for serum antibodies to herpes B virus, simian immunodeficiency virus (SIV), SRV, and simian T-lymphotropic virus type 1. The animals were fed and housed according to regulations set forth by the Guide for the Care and Use of Laboratory Animals (31) and the Animal Welfare Act at the Tulane National Primate Research Center (TNPRC). For all procedures, animals were anesthetized with 10 mg ketamine/kg of body weight. The Tulane University Institutional Animal Care and Use Committee (IACUC) approved all protocols and procedures for these studies.
Immunization.
Three different recombinant VSVs (rVSVs) were mixed and used as a vaccine. Four RMs were immunized with a mixture containing 1 × 107 PFU rVSV–SRV-2 Gag, 1 × 107 PFU rVSV–SRV-2–ENVTrunc, and 1 × 107 PFU VSV–SRV-2–EnvGCYT. Macaques were anesthetized and held in an upright position with their heads tilted back. Vaccines were administered by direct instillation of 0.25 ml into each nostril (i.n.), using a syringe barrel with no needle. The initial immunizations (60 days prior to challenge) were done with the Indiana serotype [VSV G(Ind)] and the boost (30 days prior to challenge) with the Chandipura serotype [VSV G(Ch)]. Four RMs served as controls and received the VSV vector alone.
Animal inoculation and sample collection.
All vaccinated animals and sham-vaccinated controls were challenged with SRV-2 at 2 months postimmunization. Animals were inoculated intravenously with 1 ml of plasma containing 2 × 106 vRNA copies/ml collected during primary infection of an RM infected with SRV-2. Blood was collected during and after each vaccination, biweekly for the first 2 weeks postinfection (p.i.), weekly for the next 2 weeks, bimonthly up to 2 months p.i., and monthly up to 3 months p.i. Mononuclear cells were separated from the blood using Ficoll density gradient centrifugation.
SRV-2 RNA quantification.
RNA was extracted from 280 μl of EDTA plasma by using a QIAamp viral RNA kit (Qiagen, Valencia, CA) and eluted in 60 μl of nuclease-free water. Viral RNA was quantified in a plasma real-time PCR (RT-PCR) assay specific for SRV-2. Briefly, total RNA was retrotranscribed into cDNA using a TaqMan Gold RT-PCR kit and random hexamers (PE Applied Biosystems, Foster City, CA). PCRs were carried out in a spectrofluorometric thermal cycler (ABI Prism 7700 sequence detection system; PE Applied Biosystems, CA). The quantification was based on the amplification of a 47-bp fragment located in the gag region (positions 1303 to 1348). The primers and probe were as follows: forward primer, 5′-ATTAATAAGGCATCTTCCTTTTGGAAGGT-3′; reverse primer, 5′-CAGGCAACATGGAGAGTCACT-3′; and probe, 5′–6-carboxyfluorescein (FAM)–CTGTCCGGGCCATTTG–nonfluorescent quencher (NFQ)–3′. The RT-PCR conditions were as follows: 7 mM MgCl2; 200 μM dATP, dCTP, and dGTP; 400 μM dUTP; 900 nM each primer; 300 nM the probe. The amplification protocol consisted of incubation for 2 min at 50°C, followed by 10 min at 95°C and subsequently 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. All real-time PCRs were carried out in duplicate. Absolute viral RNA copy numbers were deduced by comparing the relative signal strength to the corresponding values obtained for eight 10-fold dilutions of a standard RNA that was reverse transcribed and amplified in parallel. The target copy numbers of SRV-2 were determined by using an SRV-2 standard which was constructed as follows. A larger SRV-2 gag fragment (223 bp; positions 1143 to 1365 of the SRV-2 genome) encompassing the fragment targeted by RT-PCR was PCR amplified with primers WH1.2 (5′-CCGCTGTGATGGCGGTAGTCAATCC-3′) and WH3.2 (5′-CTGGGAAAATATCCTTGGGAGGATATTC-3′), followed by a nested PCR with primers RC14 (5′-CAATCCTAAAGAGGAACTCAAAGA-3′) and RC15 (5′-TATTCTTCTGTGTTTTTATTAATAAGG-3′). This PCR product was cloned into a pCR2.1 vector (Invitrogen), and the recombinant plasmid was transformed into Escherichia coli. The insert was sequenced to confirm the size and identity of the template. The plasmid was linearized using the HindIII enzyme, and RNA transcripts were prepared from the linearized plasmid by use of a Megascript high-yield transcription kit (Ambion). The transcripts were purified by phenol-chloroform extractions, resuspended in nuclease-free water, and quantified by a spectrophotometer at an optical density of 260 nm (OD260) by use of an extinction coefficient of 40 g/ml/OD unit. The detection limit of the SRV-2 quantification assays was 102 SRV-2 RNA copies per ml of plasma.
Antibodies and flow cytometry.
Mononuclear cells derived from peripheral blood were stained for flow-cytometric analysis using a four-color method as described earlier (8, 9, 33), with the following combinations of monoclonal antibodies (and clones): CD3 (SP34)-fluorescein isothiocyanate (FITC), CD8 (Leu2a)-phycoerythrin (PE), CD4 (L200)-peridinin chlorophyll A protein (PerCP), and CD20 (2H7)-allophycocyanin (APC); CD95 (DX2)-FITC, CD3 (SP34)-PE, CD4-PerCP, and CD28 (CD28.2)-APC; and CD3-FITC, CD8-PE, HLA-DR (L243)–PerCP, and CD4 (L200)-APC (BD Biosciences Pharmingen, San Diego, CA). Cells were incubated with an excess amount of monoclonal antibodies at 4°C for 30 min, followed by a phosphate-buffered saline (PBS) wash (400 × g; 7 min) and fixation in 2% paraformaldehyde. All Abs were validated and titrated using RM peripheral blood mononuclear cells (PBMCs). Flow-cytometric acquisition and analysis of samples were performed on at least 100,000 events with a FACSCalibur flow cytometer driven by the Cell Quest software package (Becton Dickinson). Analysis of the acquired data was performed using FlowJo software (Tree Star, Inc., Ashland, OR). Cell blood counts (CBCs) were performed for each animal and each time point and were used to determine the absolute numbers of T and B cells.
IFA assay.
Antibody titers in serum samples for the challenge virus SRV-2 were tested by using an immunofluorescent antibody (IFA) assay (Bioreliance, Rockville, MD). Sera and controls were diluted 1:10 in PBS. Twenty to 25 microliters of diluted specimens and controls was added to each well of the slide. The slides were placed at 37 ± 2°C for 30 min in a moist chamber. Specimens and controls were removed by vacuum aspiration. The slides were then washed for 5 min by immersion in a jar containing PBS. Secondary antibody conjugated to fluorescein was diluted in PBS containing 0.02% Evans blue. The slides were placed again at 37 ± 2°C for 30 min in a moist chamber. Secondary antibody was removed by vacuum aspiration. The slides were then washed for 5 min by immersion in a jar containing PBS. The slides were then dried by blotting them carefully between two paper towels. The slides were then covered with mounting buffer and coverslips. The interaction between antigen and antigen-specific serum antibodies was detected by the fluorescence of the indicators under UV light. The detection was done using an epifluorescence microscope. Serum samples found to be positive at 1:10 dilution were serially diluted.
SRV-2 neutralizing antibody assay.
The titer of serum antibody capable of SRV-2 neutralization was measured as a reduction of virus-induced cytopathic killing on Raji cells. Briefly, 100 μl of cell-free virus (20 50% tissue culture infective doses [TCID50]) grown in Raji cells was added to serial dilutions of test serum in 100 μl of growth medium in triplicate wells, making serum dilutions of 1:10 to 1:640. These mixtures were preincubated for 30 min at 4°C before the addition of 5 × 104 cells. Neutralization titers are reported as the dilution of serum required to inhibit SRV-2-induced syncytium formation and cytopathic effects (which normally occur in 6 to 9 days in the absence of neutralizing antibodies).
Statistical analysis and data presentation.
The flow cytometry parameters of the groups were analyzed for significant differences (P < 0.05) by using the Mann-Whitney test. Correlations were calculated and expressed as the Spearman coefficient of correlation.
RESULTS
Construction and recovery of VSV recombinants expressing SRV-2 Gag and Env constructs.
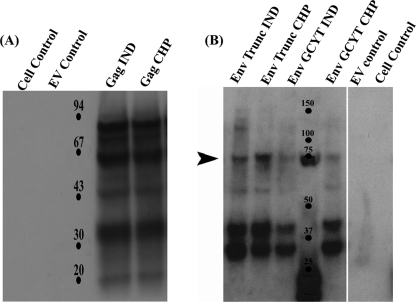
The appropriate insertions of the SRV-2 gag and env genes within the VSV genomes were confirmed by direct DNA sequencing of viral RNA after RT-PCR amplification of specific cDNA regions. BHK-21 cells were infected with the recombinant VSVs expressing Gag and Env glycoprotein constructs, and the infected cells were used to check for protein expression using IFA and Western immunoblot analysis (Fig. 2A and B), using anti-FLAG antibodies (Gag) or pooled sera from five SRV-2 positive RMs (Env constructs). The antibodies readily detected expression of the Gag and Env constructs in infected cells, while cell controls and VSV empty-vector controls produced no reaction with the antibodies.
Fig. 2.
Plaque-purified rVSV expressing SRV-2 Gag and Env construct proteins. (A) rVSV glycoprotein exchange vaccine vectors expressing SRV-2 Gag protein were assayed using Western immunoblotting. Anti-FLAG antibodies were used to detect the expression. Appropriate cell and empty-virus-vector (EV) controls were maintained. (B) rVSV glycoprotein exchange vaccine vectors expressing SRV-2 EnvTrunc proteins and EnvGCYT proteins. The expressed proteins were detected using pooled sera from five known SRV-positive monkeys at the TNPRC. Goat anti-monkey HRP conjugate was used as the secondary antibody. Indiana (IND) and Chandipura (CHP) strains of VSV were used to clone SRV-2 Gag and Env fragments.
Clinical follow-up.
All rVSV vaccinations were well tolerated, and no adverse effects were noted. After SRV challenge, none of the vaccinated RMs showed any clinical signs of SRV infection, weight loss, opportunistic infection, or increases in the sizes of lymph nodes during or after acute SRV-2 infection, and no clinical signs of AIDS were observed at the end of the follow-up, at 90 days p.i. (Table 1). Three out of the four control RMs showed significant weight loss. Two of them (DR75 and T119) developed significant anemia (average hemoglobin concentration, 2.6 g/dl) and were euthanized within 7 to 8 weeks (on day 46 p.i., T119, and on day 53 p.i., DR75). In these two RMs, in addition to low hematocrit values, lymphadenopathy, and splenomegaly occurred within 2 months after inoculation. All four RMs in the vaccinated group were alive at 3 months p.i. Vaccinated animals remained healthy, with no clinical signs of anemia and weight loss. Spleen and lymphoid tissue pathology in unvaccinated animals showed hyperplasia and provided evidence of advanced disease.
Table 1.
Clinical findings of vaccinated and control macaques following SRV-2 challenge
| Characteristic | Value for indicated animal |
|||||||
|---|---|---|---|---|---|---|---|---|
| Vaccinated |
Controla |
|||||||
| AK17 | FC55 | GN67 | GN69 | AN40 | DR75 | DV41 | T119 | |
| Clinical disease | − | − | − | − | + | + | − | + |
| Time to death (days) | Alive (90) | Alive (90) | Alive (90) | Alive (90) | Alive (90) | 53b | Alive (90) | 46b |
| Weight loss/gain (%) | 4.4 | 8.0 | 0.1 | 10.5 | −3.0 | −8.0 | 3.5 | −7.5 |
| Hemoglobin concn (g/dl) | 12.3 | 12.8 | 9.0 | 10.6 | 11.2 | 2.5 | 12.9 | 2.7 |
| Hematocrit value (%) | 37.9 | 38.3 | 30.7 | 34.1 | 37.7 | 8.0 | 39.8 | 9.5 |
| Lymphoid hyperplasia | − | − | − | − | + | + | − | + |
| Splenomegaly | − | − | − | − | + | + | − | + |
Bold values indicate below-normal levels.
Euthanized with simian AIDS virus.
Dynamics of SRV-2 replication.
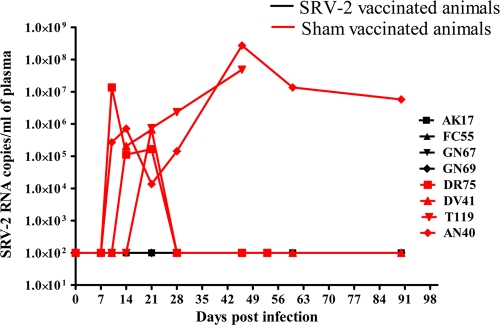
Following SRV-2 challenge, RMs were monitored for the control of plasma viral loads (VLs) as measures of vaccine efficacy. No viral replication was observed among vaccinated animals at up to 3 months postchallenge, translating into a clinical survival benefit. All four control macaques got infected and replicated the virus at 106 to 108 vRNA copies/ml of plasma. Two of the control macaques showed high-level viral replication throughout the course of infection, whereas the remaining two of four control RMs were able to drive SRV-2 replication below the detection limit from day 28 postchallenge onwards. The dynamics of plasma VLs is shown in Fig. 3. Undetectable levels of VLs in vaccinated groups were thus associated with prolonged survival of animals. In order to ascertain the presence of virus in cells of vaccinated macaques, DNA was isolated from peripheral blood cells and subjected to nested PCR using primers (WH1.2, WH3.2/RC14, and RC15) as described above. No PCR amplification of SRV DNA was observed in any of the four vaccinated animals, confirming the absence of virus at the cellular level.
Fig. 3.
Dynamics of virus replication. Plasma virus concentrations in vaccinated (black) and sham-vaccinated (red) macaques for 3 months after SRV-2 challenge.
Changes in B-cell populations.
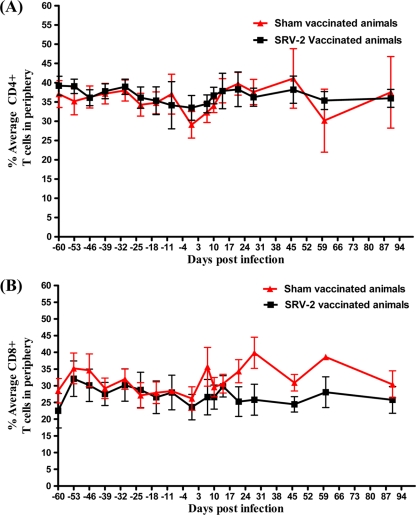
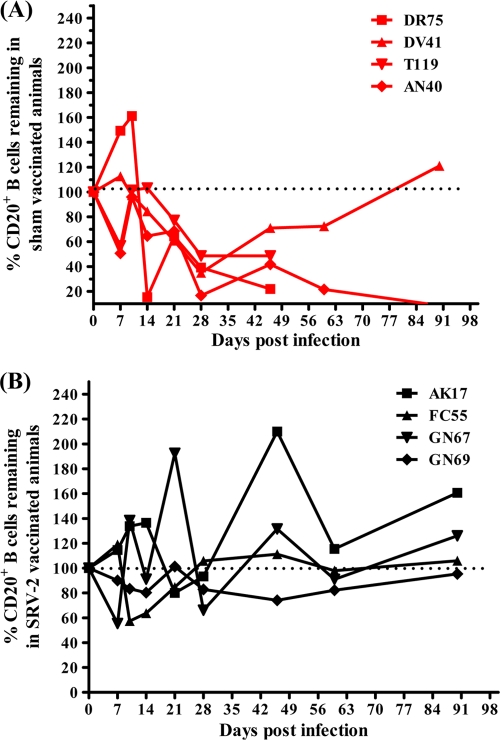
Although SRV has been shown to replicate in T cells in vivo (27, 29), immunophenotyping of immune cell populations did not show any significant changes of CD4+ lymphocytes during SRV-2 infection of RMs (Fig. 4). All macaques maintained stable peripheral-blood T-cell levels. In stark contrast, the kinetics of CD20+ B cells in blood samples of the control RMs showed that SRV-2 infection induced a significant depletion of CD20+ B cells (Fig. 5), suggesting that CD20+ B cells are the major targets supporting viral replication in SRV-2-infected RMs. In vaccinated animals, B cells were preserved throughout the study. Significant correlation was observed between survival and percentage of CD20+ B cells remaining at 3 months postchallenge.
Fig. 4.
Kinetics of T lymphocytes in vaccinated (black) and sham-vaccinated (red) macaques following SRV-2 challenge. (A) Percent average of CD3+ CD4+ T lymphocytes in peripheral blood. (B) Percent average of CD3+ CD8+ T lymphocytes in peripheral blood.
Fig. 5.
Changes in the number of circulating CD20+ B cells. The percentage of CD20+ B cells remaining were calculated for each animal by using day 0 values as 100%. (A) Percentage of B cells remaining in the four sham-vaccinated animals (red). (B) percentage of B cells remaining in the four animals vaccinated with rVSVs expressing SRV-2 Gag and Env proteins (black).
Vaccination elicits protective neutralizing Ab titer.
The ability of the sera from rVSV vaccinated and control macaques to neutralize the homologous SRV-2 in vitro was measured, and the neutralizing antibody titers correlated well with vaccine-induced protection in SRV-2 infection of macaques. Titers of neutralizing antibody against SRV-2 were assessed at three time points: prior to immunization, on the day of challenge, and on day 46 postchallenge. None of the animals had detectable SRV-2-neutralizing antibody at the time of challenge. By day 46 postchallenge, neutralizing antibody was observed only in vaccinated macaques, whose plasma RNA VLs were undetectable and ranged from 1:20 to 1:40 (Table 2). The four unvaccinated RMs did not show any neutralizing antibody activity by day 46 postchallenge (Table 2). SRV-2-specific Ab responses in longitudinal plasma samples were measured using IFA. As expected, no Ab activity was detected in samples prior to vaccination and challenge. Four of the four vaccinated animals mounted detectable antibody response as early as 28 days postchallenge, whereas only one of four controls showed delayed antibody response (see Table S2 in the supplemental material).
Table 2.
IFA and neutralizing-antibody titers for vaccinated and sham-vaccinated macaques challenged with SRV-2a
| Group and animal | Titer at day 46 postchallenge |
|
|---|---|---|
| IFA | Neutralizing antibody | |
| Control | ||
| AN40 | − | − |
| DR75 | − | − |
| DV41 | 1:40 | − |
| T119 | − | |
| Vaccinee | ||
| AK17 | 1:160 | 1:40 |
| FC55 | 1:160 | 1:40 |
| GN67 | 1:40 | 1:40 |
| GN69 | 1:80 | 1:20 |
−, none detected. No IFA or neutralizing antibodies were detected in any of the macaques prior to vaccination.
DISCUSSION
The VSV vectors that were employed in this study have shown promise as a preventive vaccine against type D simian retrovirus infection in NHPs. Because of poor efficiency in recovering rVSV with full-length SRV-2 envelope glycoprotein, SRV-2 envelope constructs were generated as secreted or as a secreted portion fused in-frame to the VSV-G cytoplasmic domain. It has previously been shown that the full-length HIV-1 Env protein was not incorporated into VSV or rabies virus unless its cytoplasmic domain was substituted with that of VSV-G (17, 28, 32). We also presumed that the G cytoplasmic tail of the VSV-GCYT construct would help in the trimerization of Env and present conformational epitopes to the immune system. Consequently, both versions of the envelope glycoproteins were used for primary vaccination and the subsequent booster. Our results indicated that vaccination with rVSV vectors expressing SRV-2 gag and env resulted in resistance to infection by challenge virus. Recombinant VSVs are known to nonspecifically incorporate certain other viral and cellular glycoproteins into their virions without adversely affecting viral infectivity (37).
It is expected that strong immune responses are generated by VSV, especially when administered via the intranasal route; therefore, in our study, RMs were vaccinated through the intranasal route using a prime-boost strategy. Upon lethal challenge with SRV-2, vaccine efficiently protected vaccinated animals and all vaccinated RMs were clinically healthy. They showed no viral replication and displayed little or no loss of CD20+ B cells. In contrast, the macaques in the control group were infected, and SRV disease onset occurred rapidly in two of four RMs. Tracking many infections in SRV-infected macaques has consistently revealed that some animals will recover and others will progress. The SRV-2 pathogenicity seen in the control group is typical of SRV infections in macaques in that one-third to one-half will recover and one-half to two-thirds will progress (7, 24).
Our study also identified an unreported feature of SRV infection, i.e., levels of CD20+ B cells are the best predictor of SRV disease. Vaccinated animals showed no destruction of B cells, in striking contrast to the control RMs, which showed severe depletion of B cells in the periphery. During acute infection, RMs from the control group lost more than 33% of their prechallenge CD20+ B cells, compared with 7% loss in vaccinated animals. By day 46 postchallenge, control macaques lost more than 54% of CD20+ B cells while vaccinated macaques gained more than 31% of CD20+ B cells compared to their prechallenge levels. CD20+ B cells rebounded in one of the four control RMs, and this recovery was associated with control of plasma VLs. The observed decreases in CD20+ B cells in all 4 control group animals may represent the early pathogenesis of SRV-2 in RMs. Future long-term studies of recovered animals will be required to determine if CD20+ B cells recover to normal preinfection levels.
SRV-2 infection showed no effect on T-cell population throughout the course of study. CD4+ T cells never showed any decline, suggesting that the receptor for SRV is a molecule other than CD4. While much is known about CD4, the receptor for HIV and its simian counterpart SIVs, and about the interaction of the receptor with these viruses, very little is known about the nature of the receptors and about the virus-receptor interaction for the majority of retroviruses, including SRV. The receptor shared by a large group of retroviruses took many years to identify. By exploiting the neutralization epitope of SRV-1, a molecule of 60 kDa was identified as a putative receptor on Raji cells (1, 41). This receptor also conferred susceptibility to infection by the type D retroviruses (40). The type D receptor is different from that used by HIV/SIV, yet both type D retroviruses and HIV/SIV cause AIDS. Identification of such receptors may help in understanding their role in inducing immunosuppression.
SRV-2-specific antibody response was seen in all the vaccinated animals as early as 2 weeks after challenge, with titers ranging from 1:10 to 1:160. It appears that an early effect on circulating B cells is an important factor in the lack of serum antibody responses and concomitant rapid disease progression.
Our results indicate that low or undetectable plasma viral load is associated with neutralizing antibody response, suggesting that the protective effect of vaccine is conferred by the presence of neutralizing antibodies. These data are consistent with previous studies, in which protective neutralizing antibodies have been induced by killed and recombinant SRV-1 and SRV-2 vaccines (14, 26). Production of neutralizing antibodies, which effectively prevents virus particles from infecting their target cells, is likely to play an important role in the response to SRV infection.
The protective role of Nabs is further suggested by the observation that one of the control RMs that did not progress to AIDS during the follow-up showed very high SRV-2-specific antibody titers (1:10 to 1:320). These observations therefore suggest that antibodies alone are also sufficient to control SRV replication during chronic infection.
In conclusion, VSV-SRV vaccination generated antibody responses that correlated with protection from detectable SRV-2 in blood and tissues. Moreover, B cells were protected from destruction by the vaccine compared to what was found for the controls. VSV-SRV vaccine appears to be a safe and promising approach for preventing an SRV-induced immunosuppressive disease in macaques. Our principal contributions were that we showed protection against SRV with a new vaccine and found that depletion of B cells, not T cells, plays a role in pathogenesis and protection of SRV-2 by the vaccine. This is a new concept for pathogenesis never before made in this model.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by NIH/NCRR base grant RR000164 to the Tulane National Primate Research Center and NIH/NCRR grant P20 RR020159 to the LSU School of Veterinary Medicine.
We also thank the animal care staff at the Tulane National Primate Research Center for the valuable help.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Benjamini E., Torres J. V., Werner L. L., Malley A. 1991. Isolation and characterization of the neutralizable epitope of simian retrovirus-1 (SRV-1) and of the cell receptor for the virus. Adv. Exp. Med. Biol. 303:71–77 [DOI] [PubMed] [Google Scholar]
- 2. Benveniste R. E., Kuller L., Roodman S. T., Hu S. L., Morton W. R. 1993. Long-term protection of macaques against high-dose type D retrovirus challenge after immunization with recombinant vaccinia virus expressing envelope glycoproteins. J. Med. Primatol. 22:74–79 [PubMed] [Google Scholar]
- 3. Brody B. A., et al. 1992. Protein of macaques against infection with simian type D retrovirus (SRV-1) by immunization with recombinant vaccinia virus expressing the envelope glycoproteins of either SRV-1 or Mason-Pfizer monkey virus (SRV-3). J. Virol. 66:3950–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brody B. A., Kimball M. G., Hunter E. 1994. Mutations within the transmembrane glycoprotein of Mason-Pfizer monkey virus: loss of SU-TM association and effects on infectivity. Virology 202:673–683 [DOI] [PubMed] [Google Scholar]
- 5. Egan M. A., et al. 2004. Immunogenicity of attenuated vesicular stomatitis virus vectors expressing HIV type 1 Env and SIV Gag proteins: comparison of intranasal and intramuscular vaccination routes. AIDS Res. Hum. Retroviruses 20:989–1004 [DOI] [PubMed] [Google Scholar]
- 6. Fernandez M., Porosnicu M., Markovic D., Barber G. N. 2002. Genetically engineered vesicular stomatitis virus in gene therapy: application for treatment of malignant disease. J. Virol. 76:895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gardner M. B., Luciw P., Lerche N., Marx P. 1988. Nonhuman primate retrovirus isolates and AIDS. Adv. Vet. Sci. Comp. Med. 32:171–226 [DOI] [PubMed] [Google Scholar]
- 8. Gaufin T., et al. 2009. Limited ability of humoral immune responses in control of viremia during infection with SIVsmmD215 strain. Blood 113:4250–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gautam R., et al. 2009. SIVrcm, a unique CCR2-tropic virus, selectively depletes memory CD4+ T cells in pigtailed macaques through expanded coreceptor usage in vivo. J. Virol. 83:7894–7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haglund K., Forman J., Krausslich H. G., Rose J. K. 2000. Expression of human immunodeficiency virus type 1 Gag protein precursor and envelope proteins from a vesicular stomatitis virus recombinant: high-level production of virus-like particles containing HIV envelope. Virology 268:112–121 [DOI] [PubMed] [Google Scholar]
- 11. Hara M., et al. 2007. Detection of SRV/D shedding in body fluids of cynomolgus macaques and comparison of partial gp70 sequences in SRV/D-T isolates. Virus Genes 35:281–288 [DOI] [PubMed] [Google Scholar]
- 12. Hara M., et al. 2005. Isolation and characterization of a new simian retrovirus type D subtype from monkeys at the Tsukuba Primate Center, Japan. Microbes Infect. 7:126–131 [DOI] [PubMed] [Google Scholar]
- 13. Hirokawa T., Boon-Chieng S., Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 14. Hu S. L., et al. 1989. Protection of macaques against simian AIDS by immunization with a recombinant vaccinia virus expressing the envelope glycoproteins of simian type D retrovirus. Proc. Natl. Acad. Sci. U. S. A. 86:7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iyer A. V., et al. 2009. Recombinant vesicular stomatitis virus-based west Nile vaccine elicits strong humoral and cellular immune responses and protects mice against lethal challenge with the virulent west Nile virus strain LSU-AR01. Vaccine 27:893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson J. E., Rodgers W., Rose J. K. 1998. A plasma membrane localization signal in the HIV-1 envelope cytoplasmic domain prevents localization at sites of vesicular stomatitis virus budding and incorporation into VSV virions. Virology 251:244–252 [DOI] [PubMed] [Google Scholar]
- 17. Johnson J. E., Schnell M. J., Buonocore L., Rose J. K. 1997. Specific targeting to CD4+ cells of recombinant vesicular stomatitis viruses encoding human immunodeficiency virus envelope proteins. J. Virol. 71:5060–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kwang H. S., et al. 1988. Simian retrovirus-D serotype 1 (SRV-1) envelope glycoproteins gp70 and gp20: expression in yeast cells and identification of specific antibodies in sera from monkeys that recovered from SRV-1 infection. J. Virol. 62:1774–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kwang H. S., et al. 1987. Viremia, antigenemia, and serum antibodies in rhesus macaques infected with simian retrovirus type 1 and their relationship to disease course. Lab. Invest. 56:591–597 [PubMed] [Google Scholar]
- 20. Lawson N. D., Stillman E. A., Whitt M. A., Rose J. K. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. U. S. A. 92:4477–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lerche N. W., Osborn K. G. 2003. Simian retrovirus infections: potential confounding variables in primate toxicology studies. Toxicol. Pathol. 31(Suppl.):103–110 [DOI] [PubMed] [Google Scholar]
- 22. Lerche N. W., et al. 2001. Evidence of infection with simian type D retrovirus in persons occupationally exposed to nonhuman primates. J. Virol. 75:1783–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lichty B. D., Power A. T., Stojdl D. F., Bell J. C. 2004. Vesicular stomatitis virus: re-inventing the bullet. Trends Mol. Med. 10:210–216 [DOI] [PubMed] [Google Scholar]
- 24. Marx P. A., et al. 1985. Isolation of a new serotype of simian acquired immune deficiency syndrome type D retrovirus from Celebes black macaques (Macaca nigra) with immune deficiency and retroperitoneal fibromatosis. J. Virol. 56:571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marx P. A., et al. 1984. Simian AIDS: isolation of a type D retrovirus and transmission of the disease. Science 223:1083–1086 [DOI] [PubMed] [Google Scholar]
- 26. Marx P. A., et al. 1986. Prevention of simian acquired immune deficiency syndrome with a formalin-inactivated type D retrovirus vaccine. J. Virol. 60:431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maul D. H., et al. 1988. Simian retrovirus D serogroup 1 has a broad cellular tropism for lymphoid and nonlymphoid cells. J. Virol. 62:1768–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mebatsion T., Conzelmann K. K. 1996. Specific infection of CD4+ target cells by recombinant rabies virus pseudotypes carrying the HIV-1 envelope spike protein. Proc. Natl. Acad. Sci. U. S. A. 93:11366–11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meyer P. R., et al. 1985. An immunopathologic evaluation of lymph nodes from monkey and man with acquired immune deficiency syndrome and related conditions. Hematol. Oncol. 3:199–210 [DOI] [PubMed] [Google Scholar]
- 30. Mitaku S., Ono M., Hirokawa T., Boon-Chieng S., Sonoyama M. 1999. Proportion of membrane proteins in proteomes of 15 single-cell organisms analyzed by the SOSUI prediction system. Biophys. Chem. 82:165–171 [DOI] [PubMed] [Google Scholar]
- 31. National Research Council 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC [Google Scholar]
- 32. Owens R. J., Rose J. K. 1993. Cytoplasmic domain requirement for incorporation of a foreign envelope protein into vesicular stomatitis virus. J. Virol. 67:360–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pandrea I. V., et al. 2007. Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J. Immunol. 179:3035–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rose J. K., Gallione C. J. 1981. Nucleotide sequences of the mRNA's encoding the vesicular stomatitis virus G and M proteins determined from cDNA clones containing the complete coding regions. J. Virol. 39:519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rose J. K., Welch W. J., Sefton B. M., Esch F. S., Ling N. C. 1980. Vesicular stomatitis virus glycoprotein is anchored in the viral membrane by a hydrophobic domain near the COOH terminus. Proc. Natl. Acad. Sci. U. S. A. 77:3884–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rose N. F., et al. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539–549 [DOI] [PubMed] [Google Scholar]
- 37. Schnell M. J., Buonocore L., Kretzschmar E., Johnson E., Rose J. K. 1996. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc. Natl. Acad. Sci. U. S. A. 93:11359–11365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schnell M. J., Buonocore L., Whitt M. A., Rose J. K. 1996. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J. Virol. 70:2318–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stojdl D. F., et al. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275 [DOI] [PubMed] [Google Scholar]
- 40. Tailor C. S., Nouri A., Zhao Y., Takeuchi Y., Kabat D. 1999. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J. Virol. 73:4470–4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Torres J. V., Werner L. L., Malley A., Benjamin E. 1991. Neutralization epitope in the envelope glycoprotein of simian retrovirus-1 (SRV-1) and identification of the virus receptor. J. Med. Primatol. 20:218–221 [PubMed] [Google Scholar]
- 42. Zao C. L., et al. 2010. The complete genome and genetic characteristics of SRV-4 isolated from cynomolgus monkeys (Macaca fascicularis). Virology 405:390–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.