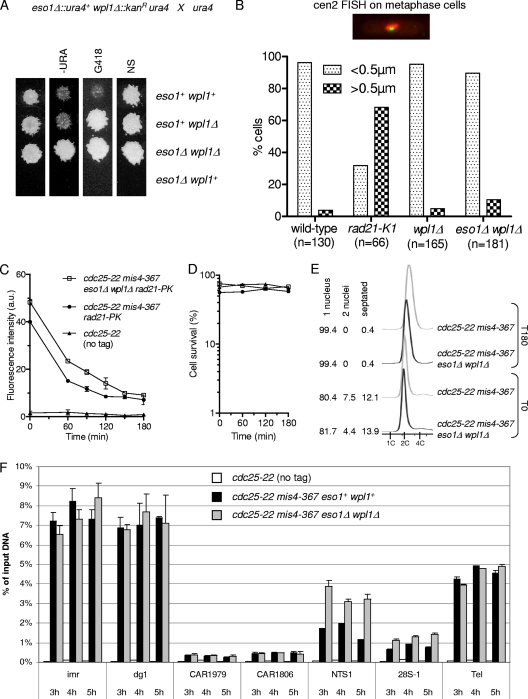
Fig. 2.
Eso1 function is dispensable when the wpl1 gene is deleted. (A) Representative example of tetrad analysis showing that eso1Δ spores do not support colony formation, whereas eso1Δ wpl1Δ spores form wild-type-size colonies. (Left) Tetrads were dissected, and spores were deposited on complete medium. Colonies were replica plated onto appropriate media to follow the segregation of eso1 and wpl1 alleles. −URA, minimum medium lacking uracil; G418, YES medium plus G418; NS, nonselective YES medium. (B) cen2 FISH on metaphase-arrested cells. Cells were arrested at metaphase by the use of the thermosensitive cut9ts mutation (anaphase-promoting complex). Fixed cells were stained for tubulin and processed for FISH using a probe mapping close to the centromere of chromosome 2 (cen2 FISH). The image shows an example of a typical metaphase cell. The mitotic spindle (red) is <2.5 μm in length, and the two cen2 FISH signals (green) are closely apposed to each other. No major defect in sister centromere cohesion was observed in wpl1Δ and wpl1Δ eso1Δ cells as opposed to the thermosensitive rad21-K1 mutant. (C to E) The stable cohesin fraction is created and is functional in eso1Δ wpl1Δ cells. Strains were cultured and processed for nuclear spreading and ChIP as in Fig. 1. (C) Kinetics of Rad21 dissociation from chromosomes. Chromatin-bound Rad21-PK was quantified from nuclear spreads. The error bars represent SD from two independent experiments. (D) Cell survival remained high throughout the duration of the experiment. (E) Cell cycle staging shows that eso1Δ wpl1Δ cells were mostly (>80%) in the G2 phase before the temperature shift and remained arrested in G2 afterward. (F) ChIP assay comparing Rad21 binding to chromosomes in eso1Δ wpl1Δ versus eso1+ wpl1+ cells from 3 h to 5 h after cohesin loading was shut off. The error bars show the SD from two quantitative PCR (qPCR) quantifications.