Abstract
Lactobacillus sanfranciscensis LSCE1 was selected as a target organism originating from recurrently refreshed sourdough to study the metabolic rerouting associated with the acid stress exposure during sourdough fermentation. In particular, the acid stress induced a metabolic shift toward overproduction of 3-methylbutanoic and 2-methylbutanoic acids accompanied by reduced sugar consumption and primary carbohydrate metabolite production. The fate of labeled leucine, the role of different nutrients and precursors, and the expression of the genes involved in branched-chain amino acid (BCAA) catabolism were evaluated at pH 3.6 and 5.8. The novel application of the program XCMS to the solid-phase microextraction-gas chromatography-mass spectrometry (SPME-GC-MS) data allowed accurate separation and quantification of 2-methylbutanoic and 3-methylbutanoic acids, generally reported as a cumulative datum. The metabolites coming from BCAA catabolism increased up to seven times under acid stress. The gene expression analysis confirmed that some genes associated with BCAA catabolism were overexpressed under acid conditions. The experiment with labeled leucine showed that 2-methylbutanoic acid originated also from leucine. While the overproduction of 3-methylbutanoic acid under acid stress can be attributed to the need to maintain redox balance, the rationale for the production of 2-methylbutanoic acid from leucine can be found in a newly proposed biosynthesis pathway leading to 2-methylbutanoic acid and 3 mol of ATP per mol of leucine. Leucine catabolism to 3-methylbutanoic and 2-methylbutanoic acids suggests that the switch from sugar to amino acid catabolism supports growth in L. sanfranciscensis in restricted environments such as sourdough characterized by acid stress and recurrent carbon starvation.
INTRODUCTION
Yeasts and lactic acid bacteria (LAB) play a key role in the generation of flavor compounds and their precursors in fermented foods. The production of alcohols, esters, aldehydes, lactones, and terpenes depends on the species involved, food system composition, and physicochemical conditions (23, 41, 51). LAB represent key organisms in sourdough fermentation: both the naturally occurring species and the inoculated ones play different roles in the dough, depending on their aptitudes and interactions with the system (6, 10, 20, 23, 41, 49). It has been reported that sourdough process parameters, including temperature, dough yield, oxygen, and pH, as well as the composition of the sourdough microbiota, determine the quality and handling properties of sourdough as well as the metabolic response of microorganisms (2, 25, 47). The exposure of microbial cells to stressful conditions during fermentation involves a broad transcriptional response with many induced or repressed genes (14, 25–27). The complex network of such responses, involving several metabolic activities, will reflect upon the composition and organoleptic properties of the dough and final products (41). Even if autochthonous bacteria are adapted and competitive, the dough can be considered a stressful environment for them (8, 30, 41). In fact, the physicochemical conditions of the habitat change during the fermentation process, mainly due to microbial acidification and nutrient breakdown (1, 8). During processing, the dynamic fluctuation of the environment itself, particularly when the dough is exposed to cyclic reactivation by inoculation of fresh raw materials with fully fermented sourdough (so-called “back slopping”), is a stress source for every microorganism involved (30).
The increased release of alcohols, ketones, keto acids, and amino acid metabolites has been observed in several LAB species following stress exposure in model systems simulating sourdough and cheese curds (15, 23, 50, 51, 55). More specifically, the exposure of different microbial species to osmotic, acid, oxidative, and chemical stresses gave rise to an accumulation (20- to 30-fold) of 3-methylbutanoic acid generated in the leucine (Leu) degradation pathway (3, 23, 34, 36, 48, 51). Systematic investigations regarding 3-methylbutanoic acid overproduction were previously performed (23, 33, 51). The interest in this behavior is 2-fold. In fact, the overproduction of a molecule implies a parallel modification in the expression of specific genes related to the stress response and to the metabolic pathways considered. Moreover, the knowledge of metabolic rerouting associated with stress supports process innovation and quality improvement and may be exploited to deliberately overproduce metabolites contributing to the flavor of fermented food (11, 41).
In this work, Lactobacillus sanfranciscensis LSCE1 was selected as a target organism because it responded with a 3-methylbutanoic acid overproduction to osmotic and acidic stress (23, 51). Moreover, L. sanfranciscensis is the main organism naturally occurring in sourdough type I, and it is usually found in starter preparations for industrial sourdough fermentations (19, 22, 24, 38, 52). Lanciotti et al. (29) reported a metabolic shift from 3-methylbutanoic acid to 2-methylbutanoic acid in several Lactobacillus spp. in milk following high-pressure homogenization stress. The mechanism of overproduction of 3-methylbutanoic or 2-methylbutanoic acid after 2 h of exposition to acid conditions was elucidated in order to evaluate the potential metabolic deviation or shift induced by stress. In particular, the fate of labeled leucine at pH 3.6 and 5.8 and the expression of the genes involved in the branched-chain amino acid (BCAA) catabolism under acid stress were investigated.
MATERIALS AND METHODS
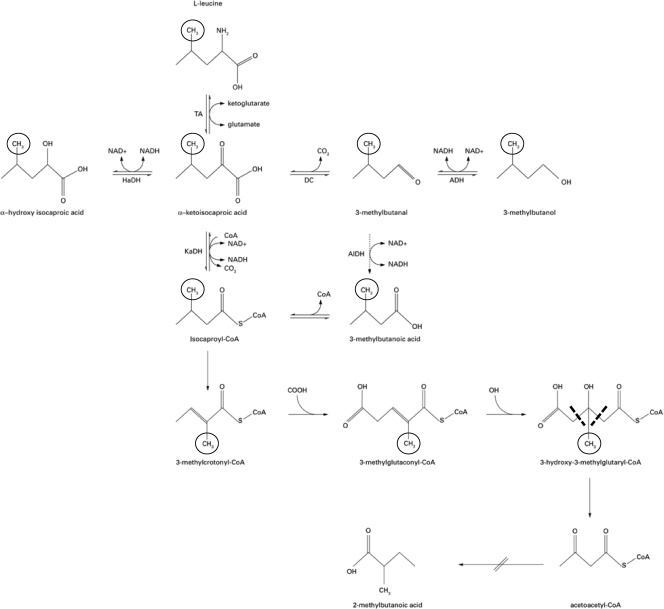
A metabolic scheme was constructed to elicit the 2-methylbutanoic and 3-methylbutanoic acid pathway on the basis of data from literature (4, 45, 46), as well as the bioinformatics pathways available on the website http://www.genome.ad.jp. The hypothetical enzymes and precursors involved in production of both compounds have been selected for the different analyses according to the proposed scheme presented in Fig. 1.
Fig. 1.
Fate of leucine in the metabolites through 3-methylbutanoic and 2-methylbutanoic acid. TA, transaminase; HaDH, hydroxy acid dehydrogenase; DC, keto acid decarboxylase; ADH, alcohol dehydrogenase; AlDH, aldehyde dehydrogenase; and KaDH, keto acid dehydrogenase. The position of the deuterium label in a methyl-group of leucine (l-leucine-5,5,5-d3) and its fate throughout the metabolic pathways to 3(3,3,3 d3)-methylbutanol and 3(3,3,3 d3)-methylbutanoic acid are indicated by a circle. The removal of the label in the branch finally leading to 2-methylbutanoic acid (abbreviated) is indicated by dashed lines. Adapted from data by Smit et al. (46).
Growth conditions.
L. sanfranciscensis LSCE1 was cultivated at 32°C in sourdough bacterium (SDB) medium (28).
For the metabolic and genetic study, the experiments were done with hydrolyzed wheat flour (WFH) medium (12), modified by the addition of leucine (Leu) or glutamate (Glut) or α-ketoglutarate (α-Ket) or citrate (Cit) (5 mM) and by the addition of maltose or fructose (Table 1). Each compound was added in separate media at a final concentration of 5 mM. The pH of the media was modified to attain the final value of 5.8 or 3.6.
Table 1.
Growth conditions of L. sanfranciscensis LSCE1
| Growth conditiona | Sugar in WFH | Precursor |
|---|---|---|
| M | Maltose | |
| M Leu | Maltose | Leucine |
| M Glut | Maltose | Glutamate |
| M α-Ket | Maltose | α-Ketoglutarate |
| M Cit | Maltose | Citrate |
| MH | Maltose (pH 3.6) | |
| MH Leu | Maltose (pH 3.6) | Leucine |
| MH Glut | Maltose (pH 3.6) | Glutamate |
| MH α-Ket | Maltose (pH 3.6) | α-Ketoglutarate |
| MH Cit | Maltose (pH 3.6) | Citrate |
| F | Fructose | |
| F Leu | Fructose | Leucine |
| F Glut | Fructose | Glutamate |
| F α-Ket | Fructose | α-Ketoglutarate |
| F Cit | Fructose | Citrate |
| FH | Fructose (pH 3.6) | |
| FH Leu | Fructose (pH 3.6) | Leucine |
| FH Glut | Fructose (pH 3.6) | Glutamate |
| FH α-Ket | Fructose (pH 3.6) | α-Ketoglutarate |
| FH Cit | Fructose (pH 3.6) | Citrate |
M, maltose; F, fructose; Leu, leucine; Glut, glutamate; α-Ket, α-ketoglutaric acid; Cit, citrate; MH and FH, maltose and fructose, respectively, at pH 3.6.
Sample preparation.
In order to adapt microbial cells to growth in WFH medium, L. sanfranciscensis LSCE1 was precultivated overnight at 32°C in 10 ml of WFH with maltose (WFH M), and cultivated (inoculum of 2% [vol/vol]) under the same conditions in 50 ml of WFH M. Finally, 300 ml of WFH M in a Bioreactor (Biostat MD; B. Braun, Germany) was inoculated with 20 ml of the overnight culture and maintained at constant temperature and pH values of 32°C and 5.8, respectively. The optical density at 560 nm (OD560) was monitored until it reached a value of 0.5 (middle exponential phase). Then, cells were harvested by centrifugation for 15 min at 6,000 rpm, washed with 200 ml of sterile water, centrifuged again, resuspended (10% vol/vol) in 200 ml of different fresh modified media (Table 1), and incubated for 2 h at 32°C. The samples at time zero (T0) were stocked immediately at −20°C (in WFH medium not inoculated [WFHTQ]). All of the samples were divided for solid-phase microextraction-gas chromatography-mass spectrometry (SPME-GC-MS), high-performance liquid chromatography (HPLC), and RNA isolation analysis. Immediately 1 ml of RNAprotect bacterial reagent (Qiagen) was added to RNA samples.
Labeled-leucine analysis.
The samples (Table 2) were prepared in a minimal medium containing vitamins, amino acids, and sugars, optimized for L. sanfranciscensis (Table 3). The final minimal medium was brought to pH 5.8 or 3.6 by using 2 M NaOH and 99% lactic acid, respectively, and the various solutions were filter sterilized (0.2-μm pore size) with a vacuum-driven disposable filtration system (Millipore Corporation, Billerica, MA). The sugars added were fructose or maltose. The precursors and the nutrients added were glutamate, 3-methylbutanal (3MBnal), and citrate at a concentration of 5 mM. The control was performed with unlabeled leucine. No other BCAAs than leucine (valine and isoleucine) were added to the synthetic media. The samples were incubated for 2 h at 32°C and stocked at −20°C until the analysis with gas chromatography-mass spectrometry (GC-MS)-SMPE by the full SCAN mode (analysis of total volatile compounds present in the sample) (i), and the select ion monitoring (SIM) mode was developed for the analysis of specific ions selected on the basis of their mass to indicate the incorporation of the labeled segment into a leucine catabolism product (39) (ii). The ion selected as the target was ion 46, and the qualifier was ion 44. Finally, a GC flame ionization detector (FID) was used for the analysis of the membrane fatty acids (iii).
Table 2.
Samples prepared for the analysis of the labeled-leucine metabolic route in a semisynthetic medium
| Sample typea | Sugar in synthetic medium | Precursor(s) |
|---|---|---|
| SM Leu | Maltose | Leucine |
| SM Leu/Cit | Maltose | Citric acid and leucine |
| SM Leu/Glut | Maltose | Glutamic acid and leucine |
| SM Leu/3MBnala | Maltose | 3-Methylbutanal and leucine |
| SM Leu (pH 3.6) | Maltose (pH 3.6) | Leucine |
| SM Leu/Cit (pH 3.6) | Maltose (pH 3.6) | Citric acid and leucine |
| SM Leu/Glut (pH 3.6) | Maltose (pH 3.6) | Glutamic acid and leucine |
| SM Leu/3MBnal (pH 3.6) | Maltose (pH 3.6) | 3-Methylbutanal and leucine |
| SM Leu+ | Maltose | Leucine+ |
| SM Leu+/Cit | Maltose | Citric acid and leucine+ |
| SM Leu+/Glut | Maltose | Glutamic acid and leucine+ |
| SM Leu+/3MBnal | Maltose | 3-Methylbutanal and leucine+ |
| SM Leu+ (pH 3.6) | Maltose (pH 3.6) | Leucine+ |
| SM Leu+/Cit (pH 3.6) | Maltose (pH 3.6) | Citric acid and leucine+ |
| SM Leu+/Glut (pH 3.6) | Maltose (pH 3.6) | Glutamic acid and leucine+ |
| SM Leu+/3MBnal (pH 3.6) | Maltose (pH 3.6) | 3-Methylbutanal and leucine+ |
| SF Leu | Fructose | Leucine |
| SF Leu/Cit | Fructose | Citric acid and leucine |
| SF Leu/Glut | Fructose | Glutamic acid and leucine |
| SF Leu/3MBnal | Fructose | 3-Methylbutanal and leucine |
| SF Leu (pH 3.6) | Fructose (pH 3.6) | Leucine |
| SF Leu/Cit (pH 3.6) | Fructose (pH 3.6) | Citric acid and leucine |
| SF Leu/Glut (pH 3.6) | Fructose (pH 3.6) | Glutamic acid and leucine |
| SF Leu/3MBnal (pH 3.6) | Fructose (pH 3.6) | 3-Methylbutanal and leucine |
| SF Leu+ | Fructose | Leucine+ |
| SF Leu+/Cit | Fructose | Citric acid and leucine+ |
| SF Leu+/Glut | Fructose | Glutamic acid and leucine+ |
| SF Leu+/3MBnal | Fructose | 3-Methylbutanal and leucine+ |
| SF Leu+ (pH 3.6) | Fructose (pH 3.6) | Leucine+ |
| SF Leu+/Cit (pH 3.6) | Fructose (pH 3.6) | Citric acid and leucine+ |
| SF Leu+/Glut (pH 3.6) | Fructose (pH 3.6) | Glutamic acid and leucine+ |
| SF Leu+/3MBnal (pH 3.6) | Fructose (pH 3.6) | 3-Methylbutanal and leucine+ |
SM, synthetic medium with maltose; SF, synthetic medium with fructose; Leu, leucine; Leu+, l-leucine-5,5,5-d3; Cit, citrate; Glut, glutamate; 3MBnal, 3-methylbutanal.
Table 3.
Synthetic medium with composition optimized for L. sanfranciscensis
| Compound | Quantity |
|---|---|
| Base solution (800 ml) | |
| K2HPO4 | 2 g |
| KH2PO4 | 2 g |
| MgSO4·7H2O | 0.2 g |
| MnSO4·4H2O | 0.04 g |
| FeSO4·7H2O | 0.02 g |
| NaCl | 0.02 g |
| Fructose or maltose | 10.53 g |
| Adenine | 0.02 g |
| Uracil | 0.02 g |
| Sodium acetate | 10 g |
| Ammonium chloride | 3 g |
| Water | 800 ml |
| Vitamin mix (100 ml) | |
| Thiamine HCl | 0.02 g/liter |
| Biotin | 0.0001 g/liter |
| Nicotinic acid | 0.05 g/liter |
| Riboflavin | 0.01 g/liter |
| Calcium pantothenate | 0.01 g/liter |
| Pyridoxal HCl | 0.02 g/liter |
| Folic acid | 0.001 g/liter |
| Water | 1 liter |
| Amino acid mix (100 ml) | |
| l-Lysine HCl, l-histidine HCl, | |
| l-arginine HCl, aspartic acid, | |
| l-threonine, l-serine, l-proline, | |
| glycine, dl-alanine, l-methionine, | |
| l-tyrosine, l-tryptophan, | |
| l-phenylalanine, and l-cysteine HCl | 0.5 g each |
| l-Leucine | 5 mM (final concn) |
| Alternatively added precursors | |
| Glutamate | 5 mM (final concn) |
| 3-Methylbutanal | 5 mM (final concn) |
| Citrate | 5 mM (final concn) |
| Water | 1 liter |
RNA extraction, reverse transcription, and gene expression.
RNA was extracted as described in reference 43. Once RNA integrity had been verified, 24-μl RNA samples were treated with 7 U DNase (Promega, Madison, WI) in 4 μl DNase buffer (Promega) and incubated at 37°C for 1 h before the reaction was stopped with 4 μl of stop solution (Promega) and incubation at 65°C for 10 min. The RNA concentration and purity were optically determined with a GeneQuant RNA/DNA calculator (Amersham Pharmacia Biotech, Piscataway, NJ). Based upon the final concentrations, 1 μg of RNA was used to rewrite the contained mRNA into cDNA by using 2 μg of random primers (Promega), 40 μM deoxynucleoside triphosphates (dNTPs) (Qbiogene), 4 μl reverse transcription (RT) buffer (Promega), 0.5 U Moloney murine leukemia virus (MMLV) reverse transcriptase, and an RNase H− point mutant (Promega) in a total volume to 20 μl. mRNA samples were also prepared without reverse transcriptase as a control for DNA contamination. The sequences of the primers used as the housekeeping gene (phosphoketolase gene) for L. sanfranciscensis were as follows: Pk-Ls1_for, 5′-GCT GGA TCA TGG TTC TCA AGC-3′; and Pk-Ls1_rev, 5′-CCT TCG AAG TAC TTA CGT AAG TC-3′. The primers were designed in this study on the basis of L. sanfranciscensis TMW1.304 genome sequences. The genes selected were identified to have an important role in isovaleric acid metabolic pathway. Other genes such as those coding for glutamate dehydrogenase, α-acetolactate decarboxylase, and CodY (a GTP-binding repressor) were tested but not found in the genome of L. sanfranciscensis LSCE1 (data not shown). The gene encoding the branched-chain amino acid transferase (bcaT; accession no. FN908479) was amplified with the primers bcAT_for (5′-CTG TCA TCT ACG TGT TAT TGA TTG-3′) and bcAT_rev (5′-GTA TAA TGG CGC ATG ATG ATG G-3′). The gene encoding the hydroxyisocaproate dehydrogenase (hid; accession no. FN908480) was amplified with the primers hid_for (5′-CAT GAT GAG ACC GTT GAC TTA G-3′) and hid_rev (5′-GTA TAA TGG CGC ATG ATG ATG G-3′). The gene encoding the glutamine aminotransferase (gluAT) (accession no. FN908481) was amplified with the primers gluAT_for (5′-GAA TAA CTG CCA ACG TTA ATC TTG-3′) and gluAT_rev (5′-GTT GGT ACA ACA GGA ATG AC-3′); the gene encoding the heat shock protein (dnaK) (accession no. FN908478) was amplified with the primers dnaK_for (5′-GGT GAA GAA GAA TAC ACA GTT-3′) and dnaK_rev (5′-CCA AGG TAA TCT TCA GCA TAA CC-3′).
The level of gene expression was determined fluorometrically with SYBR green and the Lightcycler system (Roche Diagnostics, GmbH, Mannheim, Germany). Samples were examined for differences in gene expression by relative quantification (17, 37).
GC-MS-SPME.
The GC-MS-SPME analyses were performed by a method previously developed (34), modified as follows. The samples were equilibrated for 10 min at 50°C. The SPME fiber was exposed to each sample for 40 min, and finally the fiber was inserted into the injection port of the gas chromatograph for 5 min of sample desorption. These optimal conditions were chosen on the basis of previous experiments with pure compounds analyzed after different times (10, 20, 30, 40, or 50 min) and at different temperatures (45, 50, 60, 70, or 75°C). 4-Methyl-2-pentanol was used as an internal standard at a final concentration of 200 ppm.
All of the GC-MS raw files were converted to netCDF format via Chemstation (Agilent Technologies) and subsequently processed with the XCMS toolbox (http://metlin.scripps.edu/download/). XCMS software allows automatic and simultaneous retention time alignment, matched filtration, peak detection, and peak matching. The resulting table containing information such as peak index (retention time-m/z pair) and normalized peak area was exported into R (www.r-project.org) for subsequent statistical or multivariate analyses.
Cell lipid extraction.
Cell lipid extraction was performed according to a method previously described (32). Cellular fatty acid extraction and methylation were performed with the Microbial Identification System (MIS) protocol produced by Microbial ID (MIDI, Newark, DE), also described by Welch (54).
Gas chromatographic analyses of cell FAs.
The hexane extracts of cell fatty acid methyl esters were analyzed for the identification and detection of the cell fatty acids (FAs) (21). A GC-flame ionization detector (FID) (PerkinElmer Clarus 500; PerkinElmer Life and Analytical Sciences, Shelton, CT) and a capillary column (Supelcowax-10; 30 m by 0.32 mm by 0.25 μm) were used for the analysis. The injector and the detector were both held at 240°C. The temperature was programmed from 50°C (held for 1 min) to 220°C at a rate of 4°C/min and held at 220°C for 10 min. The carrier gas was helium, with a rate of 1 ml/min, and the split was 1:10. Membrane FAs, free fatty acids (FFAs), and hydrocarbons were quantified with internal standards and identified by comparing their retention times with those of the standards mix Fame 37 and BAME mix (Supelco, Sigma-Aldrich, Milan, Italy).
Statistical analysis.
Three independent replicates were performed for all experiments. The data exposed or used for multivariate analyses are the means of the three repetitions.
The quantitative data obtained from metabolites and gene expression determinations were used to build up a single matrix, which was submitted to a two-way hierarchical clustering analysis (16). A heat map, visualizing metabolite concentration and gene expression changes, was then obtained in which values are represented by cells colored according to the Z-scores, where Z = (observed value − mean)/standard deviation.
RESULTS
Effects of stress exposure on metabolite release.
Mid-exponential-phase cells were suspended in fresh WFH containing different carbon sources (maltose or fructose) and precursors (leucine, glutamate, α-ketoglutaric acid, and citrate) and exposed for 2 h to pH 5.8 (control) or 3.6 (acid condition). No change in cell viability, detected by plate counting (data not shown), was observed. The quantitatively most important compounds released and those showing the greatest changes in relation to the control conditions after 2 h of acid stress are reported in Tables 4 and 5.
Table 4.
Volatile compounds produced by L. sanfranciscensis LSCE1 in WFH with maltose
| Metabolite | Production (mg/liter) ina: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| WFHTQ | M | MH | M Leu | MH Leu | M Glut | MH Glut | M α-Ket | MH α-Ket | |
| 2-Butanone | 1.6 | 6.6 | 16.2 | 2.1 | 7.7 | 2.1 | 2.3 | 7.6 | 1.9 |
| 3-Methyl-1-butanol | 0.8 | 4.6 | 4.8 | 4.4 | 3.6 | 2.3 | 2.0 | 3.9 | 2.2 |
| Butanoic acid | 0.1 | 3.0 | 7.0 | 4.0 | 7.5 | 2.6 | 8.1 | 5.6 | 8.7 |
| 3-Methylbutanoic acid | 0.3 | 15.8 | 54.2 | 31.5 | 71.5 | 15.0 | 82.3 | 39.6 | 87.8 |
| 2-Methylbutanoic acid | NDb | 8.1 | 25.1 | 16.7 | 33.5 | 8.1 | 41.8 | 19.1 | 44.4 |
| Phenylethanol | 0.6 | 34.7 | 20.6 | 52.4 | 28.6 | 39.3 | 40.4 | 30.6 | 39.5 |
| Total | 9.1 | 221.3 | 304.9 | 282.7 | 298.7 | 230.0 | 367.1 | 252.1 | 393.9 |
WFHTQ, hydrolyzed wheat flour medium not inoculated; M, maltose; MH, maltose at pH 3.6; Leu, leucine; Glut, glutamic acid; α-Ket, α-ketoglutaric acid. The data are means of three independent experiments. The coefficients of variability were lower than 4%.
ND, not detectable.
Table 5.
Volatile compounds produced by L. sanfranciscensis LSCE1 in WHF with fructose at pH 5.8 and 3.6
| Metabolite | Production (mg/liter) ina: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| F | FH | F Leu | FH Leu | F Glut | FH Glut | F α-Ket | FH α-Ket | F Cit | FH Cit | |
| 2-Butanone | 2.7 | 13.3 | 1.4 | 7.2 | 9.5 | 6.9 | 6.8 | 7.4 | 2.5 | 3.0 |
| 3-Methyl-1-butanol | 2.6 | 4.4 | 2.8 | 3.8 | 3.6 | 3.8 | 3.9 | 3.8 | 3.3 | 2.4 |
| Butanoic acid | 1.9 | 8.3 | 3.6 | 7.6 | 4.2 | 6.9 | 5.7 | 7.4 | 7.1 | 10.4 |
| 3-Methylbutanoic acid | 15.9 | 109.5 | 41.4 | 116.6 | 25.0 | 105.3 | 82.6 | 119.3 | 109.3 | 138.5 |
| 2-Methylbutanoic acid | 7.3 | 52.8 | 22.5 | 54.1 | 12.0 | 50.7 | 40. | 57.0 | 58.9 | 73.0 |
| Phenylethanol | 25.3 | 47.6 | 70.0 | 42.8 | 48.5 | 45.1 | 49.2 | 44.5 | 67.3 | 62.4 |
| Total | 151.0 | 424.3 | 315.6 | 400.2 | 263.5 | 387.4 | 355.3 | 414.3 | 449.4 | 491.9 |
F, fructose; FH, fructose at pH 3.6; Leu, leucine; Glut, glutamic acid; α-Ket, α-ketoglutaric acid; Cit, citrate. The data are means of three independent experiments. The coefficients of variability were lower than 4%.
The total release of metabolites, calculated by summarizing the different compounds, was strongly dependent on the conditions and increased up to 2-fold following acid stress, particularly in the presence of fructose. The molecules released under the various conditions were as follows: organic acids, short-chain fatty acids (SCFAs) such as butanoic acid, ethyl hexanoate (ranging from 1.7 to 2.4 mg/liter), octanoic acid (ranging from 1 to 9 mg/liter), 2-methylbutanoic acid, 3-methylbutanoic acid, alcohols, esters, aldheydes, and ketones. The most important molecules released under acid stress were 2-methylbutanoic acid and 3-methylbutanoic acid, which attained mean levels of 73.6 and 138.57 mg/liter, respectively. The analysis of the GC data, with the program XCMS, which identified the molecules on the basis of their ions, allowed an enhanced separation of metabolites and in particular the separation and quantification of 2-methylbutanoic and 3-methylbutanoic acids.
Tables 6 and 7 report the levels of carbohydrates, individually occurring or added to WFH, and the concentration of the organic acids released.
Table 6.
Consumption and formation of carbohydrates, ethanol, and organic acids naturally occurring or added to WFH with maltose upon 2 h of exposure of L. sanfranciscensis LSCE1 to acid stress
| Sugar, organic acid, or ethanol | Consumption or formation (mg/liter) ina: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M | MH | M Leu | MH Leu | M Glut | MH Glut | M α-Ket | MH α-Ket | M Cit | MH Cit | |
| Sugars | ||||||||||
| Maltose | −3,618 | −5,701 | −3,560 | −869 | −4,919 | −894 | 2,936 | 170 | 2,158 | 141 |
| Fructose | −200 | −236 | −175 | −233 | −254 | −189 | −265 | −199 | −235 | −176 |
| Mannitol | 141 | 1,968 | NDb | ND | ND | ND | ND | ND | ND | ND |
| Glucose | −1470 | −104 | −779 | 68 | −1,877 | 136 | 176 | 59 | 18 | 130 |
| Ethanol or organic acid | ||||||||||
| Ethanol | 560 | 1,558 | 803 | −25 | 633 | −55 | 509 | −16 | 204 | 1 |
| Acetate | 18 | 47 | 4 | 47 | 89 | 5 | 47 | −19 | 349 | −2 |
| Lactate | 971 | −67 | 4,438 | 1,184 | 1,754 | −5 | −149 | 887 | 1,124 | −188 |
M, maltose; MH, maltose at pH 3.6; Leu, leucine; Glut, glutamate; α-Ket, α-ketoglutaric acid; Cit, citrate. The data are means of three independent experiments. The coefficients of variability were lower than 6%.
ND, not detectable.
Table 7.
Consumption and formation of carbohydrates, ethanol, and organic acids naturally occurring or added to WFH with fructose upon 2 h of exposure of L. sanfranciscensis LSCE1 to acid stress
| Sugar, organic acid, or ethanol | Consumption or formation of compound (mg/liter) ina: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| F | FH | F Leu | FH Leu | F Glut | FH Glut | F α-Ket | FH α-Ket | F Cit | FH Cit | |
| Sugars | ||||||||||
| Maltose | −2,189 | −3,205 | −2,147 | −1,039 | −2,394 | −63 | −1,807 | −950 | −1,891 | −1,180 |
| Fructose | −2,048 | −2,248 | −2,058 | 170 | −2,121 | −2,273 | −1,190 | 98 | −1,713 | −112 |
| Mannitol | −142 | −312 | 481 | −80 | −1,000 | 389 | 425 | 326 | 441 | 530 |
| Glucose | −1,145 | −1,864 | −510 | 1,309 | −1,136 | −1,506 | 454 | 1,619 | 483 | 1,270 |
| Ethanol or organic acid | ||||||||||
| Ethanol | 392 | −403 | 437 | 39 | 359 | −167 | 35 | 34 | 75 | 15 |
| Acetate | 331 | −104 | 394 | −30 | 308 | −139 | 360 | −68 | 263 | 75 |
| Lactate | 1,349 | 640 | −131 | 615 | 1,523 | 94 | −133 | 801 | 1,324 | 1 |
F, fructose; FH, fructose at pH 3.6; Leu, leucine; Glut, glutamate; α-Ket, α-ketoglutaric acid; Cit, citrate.The data are means of three independent experiments. The coefficients of variability were lower than 6%.
It is important to outline that, in general, the stress exposure reduced sugar consumption, particularly that of glucose and maltose, and primary metabolite production (ethanol and acetic acid but not lactic acid). However, in the absence of precursors or nutrients, and particularly with maltose at pH 3.6 (MH), the total uptake of maltose, accompanied by a higher concentration of ethanol, was observed with respect to the controls at pH 5.8. On the other hand in the condition FH (with fructose at pH 3.6) the total sugar uptake was not accompanied by the production of ethanol, lactic, and acetic acids.
It can be observed that, despite the fact that the carbohydrates' fermentation products (ethanol, lactic acid, and acetic acid) tended to decrease with sublethal acid stress, the metabolites associated with amino acid catabolism, such as 2-methylbutanoic and 3-methylbutanoic acids, here occurred at a 1:2 ratio. However, also under the control conditions, at pH 5.8, citrate with both maltose and fructose (M Cit and F Cit, respectively), α-ketoglutarate with both maltose and fructose (M α-Ket and F α-Ket, respectively), and leucine with both maltose and fructose (M Leu and F Leu, respectively) induced a significant increase in 3-methylbutanoic and 2-methylbutanoic acids (Tables 4 and 5).
Determination of the genes involved in leucine and isoleucine degradation (PCR screening).
The genes putatively involved in the breakdown of leucine, a precursor of 3-methylbutanoic acid, and isoleucine, a precursor of 2-methylbutanoic acid, have to be considered since data are lacking regarding the total genome of L. sanfranciscensis. Moreover, highly conserved sequences of the genes involved in the branched-chain amino acid catabolism were not found specifically in the strain L. sanfranciscensis LSCE1. The selected genes were those coding for branched-chain aminotransferase (bcAT), hydroxyisocaproate dehydrogenase (hid), glutamine aminotransferase (gluAT), and a heat shock protein (dnaK).
In order to assess the impact of acid stress on the expression of genes related to leucine catabolism, the relative gene expression (17, 37) was calculated, considering as a control the unstressed samples. The genes considered were gluAT, hid, and bcAT.
The data obtained (Fig. 2) revealed that gluAT was overexpressed under acid conditions only with maltose (M) and in the presence of leucine (M Leu), glutamate M Glut), and citrate (M Cit), as well as with fructose (F). Regarding the hid gene, overexpression occurred in the M Leu, M Glut, M Cit, F, F α-Ket, and F Cit samples. The results of bcAT gene expression revealed that this gene was activated by acid stress in the presence of maltose with both leucine and glutamate. However, under the other conditions analyzed, its expression was repressed.
Fig. 2.
Relative gene expression (rge) calculated with the samples at pH 5.8 as a control and samples at pH 3.6 representing acid stress. The gene products are as follows: bcAT, branched-chain aminotransferase; gluAT, glutamine aminotransferase; hid, hydroxyisocaproate dehydrogenase. M, maltose; F, fructose; Leu, leucine; Glut, glutamate; α-k, α-ketoglutaric acid; and Cit, citrate.
Fate of labeled and unlabeled leucines.
In order to evaluate if 2-methylbutanoic acid originates from leucine catabolism and not only from isoleucine catabolism, the strain L. sanfranciscensis LSCE1 was exposed for 2 h in semisynthetic medium containing leucine as the only branched-amino acid source at both pH 5.8 and 3.6.
Table 8 reveals that 2-methylbutanoic acid was released in the presence of leucine only under acid stress, confirming that under this condition it can derive from leucine. However, the ratio of 2-methylbutanoic acid to 3-methylbutanoic acid was lower than that observed in Tables 4 and 5 relative to a nonsynthetic medium and presumably also containing isoleucine.
Table 8.
Production of 2-methylbutanoic acid and 3-methylbutanoic acid by L. sanfranciscensis LSCE1a
| Sample conditionb | Production (mg/liter) of: |
|
|---|---|---|
| 2-Methylbutanoic acid (ion 74) | 3-Methylbutanoic acid (ion 60) | |
| SM Leu | NDb | 5.3 |
| SM Leu/Glut | ND | 3 |
| SM Leu/3MBnal | ND | 3.6 |
| SM Leu/Cit | ND | 3.3 |
| SF Leu | 2.0 | 3.1 |
| SF Leu/Glut | ND | 3.1 |
| SF Leu/3MBnal | ND | 4.1 |
| SF Leu/Cit | ND | 3.75 |
| SMH Leu | 2.5 | 3.8 |
| SMH Leu/Glut | ND | 4 |
| SMH Leu/3MBnal | ND | 4.2 |
| SMH Leu/Cit | ND | 3 |
| SFH Leu | 3 | 4.2 |
| SFH Leu/Glut | ND | 4 |
| SFH Leu/3MBnal | ND | 4.3 |
| SFH Leu/Cit | 3 | 3.5 |
Production of 2-methylbutanoic acid and 3-methylbutanoic acid was calculated based on the log areas of their characteristic ions 74 and 60, respectively. Shown are results from production in semisynthetic media with maltose (SM) and fructose (SF) at pH 5.8 and 3.6 (SMH and SFH, respectively). Leu, unlabeled leucine; Glut, glutamic acid; 3Mbnal, 3-methylbutanal; Cit, citrate. The data are means of three independent experiments. The coefficients of variability were lower than 4%.
ND, not detectable.
The strain was exposed (8 log CFU/ml) for 2 h in a synthetic medium containing labeled (l-leucine-5,5,5-d3) or unlabeled leucine. Since only a few genes associated with leucine catabolism were found, only l-leucine-5,5,5-d3 was adopted to study the possible shift in the BCAAs' breakdown. The analytical approach used detects and quantifies only the metabolites incorporating the labeled segment, which is indicated in Fig. 1 (3-methylbutanol, 3-methylbutanal, and 3-methybutanoic acid). The figure illustrates the fate of the labeled segment. No labeled ions which can be attributed to labeled 2-methylbutanoic acid were found.
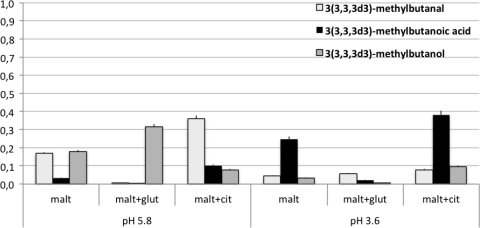
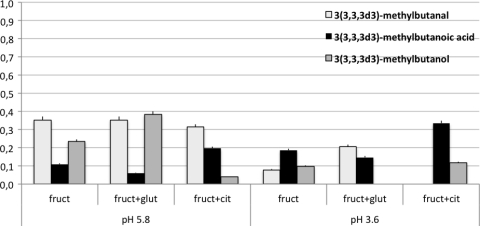
The analysis of the selected ions revealed that the labeled segment, derived from l-leucine-5,5,5-d3, was present in 3-methylbutanoic acid particularly at pH 3.6. On the other hand, labeled 3-methylbutanol and 3-methylbutanal concentrations were higher in all of the controls at pH 5.8 (Fig. 3 and 4). These data confirm that the proportion of l-leucine-5,5,5-d3 transformed into 3(3,3,3d3)-methylbutanoic acid, instead of 3(3,3,3d3)-methylbutanol and 3(3,3,3d3)-methylbutanal, was significantly higher under acid conditions. In fact, the increase in labeled 3-methylbutanoic acid was accompanied by a decrease in 3(3,3,3d3)-methylbutanal and 3(3,3,3d3)-methylbutanol. The comparison of SIM and SCAN analyses indicated that 2-methylbutanoic acid was present but not in the labeled form.
Fig. 3.
Amount of labeled compounds derived from l-leucine-5,5,5-d3 in the presence of maltose (malt) and acidic stress (in mg/liter, calculated on the basis of the log area of the target labeled ion). glut, glutamate; cit, citrate.
Fig. 4.
Amount of labeled compounds derived from l-leucine-5,5,5-d3 in the presence of fructose (fruct) and acidic stress (in mg/liter, calculated on the basis of the log area of the target labeled ion). glut, glutamate; cit, citrate.
BCAAs, 2-methylbutanoic acid, and 3-methylbutanoic acid are precursors also of iso and anteiso membrane fatty acids. The fatty acid composition of the cells exposed under the various conditions was detected (data not shown). The analysis revealed that, at least under these conditions, in L. sanfranciscensis LSCE1 BCFAs were not present.
DISCUSSION
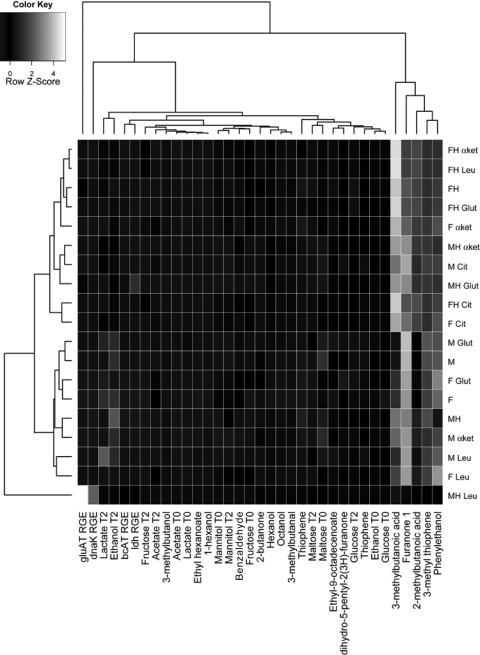
A multivariate analysis using a heat map was performed in order to identify the molecules or the genes able to significantly contribute to the statistical discrimination between stressed and unstressed conditions. Two main clusters were obtained on the basis of all of the variables. They grouped separately the stressed and unstressed samples (Fig. 5). The condition MH Leu clustered alone. The heat map underlined the role of 2-methylbutanoic and 3-methylbutanoic acids in grouping the samples under stressed and unstressed conditions. However, F α-ket, M Cit, and F Cit clustered with the stressed samples. On the other hand MH clustered with unstressed samples. A strong contribution to the clustering was also provided by lactic acid and ethanol associated with the nonstressed sample cluster. The relative gene expression of gluAT and dnaK specifically generated the separation of the sample MH Leu.
Fig. 5.
Heat map of correlations between metabolites produced by L. sanfranciscensis LSCE1 and the acid stress and nonstress conditions tested. Each square represents the Spearman's correlation.
Current literature reports the overproduction of 3-methylbutanoic acid by LAB as a consequence of stress exposure and its occurrence in several fermented foods, including cheeses and sourdough, etc. (5, 23, 42, 46, 51, 56). However, frequently cumulative values of 3-methylbutanoic and 2-methylbutanoic acids are reported as 3-methylbutanoic acid due to the difficulty of separating these compounds with the current GC techniques. In this study, the analysis of the GC data with the program XCMS, which identifies the molecules on the basis of their ions, permitted precise separation of metabolites and, in particular, accurate separation and quantification of 2-methylbutanoic and 3-methylbutanoic acids. This new opportunity revealed that under acid conditions, L. sanfranciscensis LSCE1 overproduces both 3-methylbutanoic and 2-methylbutanoic acids, which generally occur in a 2:1 ratio. Under the acid conditions (pH 3.6) and in the presence of specific nutrients, these molecules, particularly 3-methylbutanoic acid, were the predominant metabolites, among those detected by GC-MS-SPME, released after 2 h of acid stress exposure. The release of the two molecules was generally higher in the presence of fructose than in the presence of maltose. This can be explained by fructose as an electron acceptor enabling regeneration of NAD needed for dehydrogenation both of α-ketoisocaproic acid or 3-methylbutanal to enable formation of 3-methylbutanoic acid. The acid stress implied less carbohydrate utilization and ethanol, lactate, and acetate production, with the exception of MH and FH conditions. This first suggests that the cells try to minimize acid stress by limiting lactate production in favor of sugar alcohols (mannitol) or acids with higher pK values. In turn, the NAD pool, required for glycolysis and for 2-methylbutanoic or 3-methylbutanoic acids, needs regeneration, which is carried out via fructose reduction. In the stage of lactate production in the presence of fructose (without acid stress), utilization of fructose enables acetate production and ATP formation in the acetate kinase reaction. Under acid stress, this option is limited since fructose is required as a prior electron acceptor in order to achieve glycolysis with reduced lactate formation. A similar effect is seen from citrate, which also can act as electron acceptor and further enhances formation of 3-methylbutanoic acid. Under acid conditions, citrate is protonated to a higher extent and easily penetrates the cell membrane without any transport protein required. As a consequence, the production of these compounds is further enhanced. This demonstrates that the availability of electron acceptors is a clear limit for their production. Another limit is set by the availability of α-ketoglutarate for the transamination reaction. As expected, this cosubstrate enhances the formation of 2-methylbutanoic and 3-methylbutanoic acids. As for citrate, protonation at low pH apparently enhances its availability for the transamination reaction. The role of glutamate would be expected through its conversion to γ-aminobutyric acid, which is transported out of the cell via the glutamate antiporter contributing to proton motive force generation. The ammonium generated in this reaction would further contribute to compensate for pH stress. In this case, the enhanced formation of 3-methylbutanoic acid and ammonium in the presence of glutamate would be explained by its contribution to intracellular pH stabilization enabling a generally improved metabolism rather than its direct involvement as a cosubstrate.
The acid stress generated a shift in the labeled leucine products released. In fact, under all the conditions, and particularly in the presence of fructose, the labeled 3-methylbutanoic acid concentration increased and in parallel the labeled 3-methylbutanol and 3-methylbutanal decreased with respect to the corresponding controls at pH 5.8 (Fig. 3 and 4). This behavior was particularly evident in the presence of citrate, as reported above. The physiological effect of the conversion of leucine to 3-methylbutanal and further to 3-methylbutanol is the regeneration of NAD. On the contrary, the conversion of leucine to 3-methylbutanoic acid is more convenient from the energetic point of view under stress conditions, because it generates a mol of ATP per mol of leucine.
The gene expression analysis confirmed that some genes associated with the BCAAs' catabolism, such as bcAT and hid, were overexpressed under acid conditions, above all in the presence of maltose, leucine, and glutamic acid. In particular, gluAT was overexpressed under all of the acid stress conditions, presumably due to its relationship with the glutamate decarboxylase (GAD) associated with the mechanism of cellular homeostasis activated by acid stress (7, 35, 40, 41, 44).
Under the adopted experimental conditions, which did not produce any decrease in viability of L. sanfranciscensis LSCE1, the acid stress, within 2 h, was accompanied by a reduction of the carbohydrate metabolism, as shown by the decrease of ethanol, acetate, and lactate and always by a relevant increase in both 3-methylbutanoic acid and 2-methylbutanoic acid, particularly in the presence of fructose. This result was confirmed at least for 3-methylbutanoic acid when labeled leucine was supplemented as the exclusive BCAA. However, in the condition FH, not supplemented with precursors and nutrients, a total consumption of fructose, maltose, and glucose was not accompanied by ethanol and acetate production.
The overproduction of 3-methylbutanoic acid during acid stress can be attributed to the need to maintain the redox balance. In fact, under acid conditions, the principal response mechanism engages proton expulsion by FoF1 ATPase (7, 9, 47). In Staphylococcus aureus, the gene nuoF, encoding an NADH dehydrogenase, was found to be upregulated. This dehydrogenase moves 2H+ across the membrane and out of the cells, converting one NADH to NAD+, and thereby helps to increase the internal pH in the cells (4). This mechanism can activate the 3-methylbutanoic acid synthesis, which is NAD+ dependent and produces NADH during the reaction (53), therefore maintaining the NAD/NADH balance in L. sanfranciscensis LSCE1 in the presence of acid stress. This hypothesis was confirmed by hid expression under the acid stress condition. In fact, the expression of hid is NAD dependent and generates NADH from α-hydroxyisocaproic acid to α-ketoisocaproic acid. The overexpression of this gene is probably due to its NAD-dependent nature.
Concerning 2-methylbutanoic acid, it is known that generally it is produced from isoleucine (46). However, according to the results obtained, in semisynthetic media with leucine added as the only source of BCAAs, and only in the presence of acid stress, leucine resulted in being a potential precursor of 2-methylbutanoic acid also and not only of 3-methylbutanoic acid. The rationale behind the production of 2-methylbutanoic acid from leucine under stress conditions in L. sanfranciscensis LSCE1 can be found in the new biosynthesis pathway regarding the biosynthesis of 2-methylbutanoic acid from leucine in L. lactis (18). Ganesan et al. (18) showed that, during carbon starvation and onset of nonculturability, L. lactis is capable of consuming leucine via a long metabolic route to produce ATP. This route, which is initiated by leucine transamination to α-ketoisocaproic acid being subsequently oxidatively decarboxylated to isovaleryl-coenzyme A (CoA), leads to the production of 2-methylbutanoic acid and 3 mol of ATP per mol of leucine. As reported above, the best known pathway from leucine to 3-methylbutanoic acid produces 1 mol of ATP per mol of amino acid. Gene expression profiling and genome sequence analysis revealed that lactococci contain redundant genes for BCFA production, which was regulated by unknown mechanisms linked to carbon metabolism. Moreover, the new pathway to 2-methylbutanoic acid from leucine proposed in reference 18 can account for the absence of labeled 2-methylbutanoic acid in the semisynthetic medium supplemented with labeled leucine in L. sanfranciscensis LSCE1. In fact, in the presence of l-leucine-5,5,5-d3, the labeled segment would be removed in the step from 3-hydroxy-3-methyl-glutaryl-CoA to acetoacetyl-CoA (18). Therefore, according to this metabolic pathway, unlabeled 2-methylbutanoic acid would be generated from l-leucine-5,5,5-d3.
It can be assumed that the 2-methylbutanoic acid synthesis from leucine can compensate for an energy requirement. In fact, an interesting feature of leucine conversion to 2-methylbutanoic acid is the molecular efficiency in the production and use of intermediates and cofactors resulting in energy (3 mol ATP) and reducing equivalents. This phenomenon contributes to the microorganisms' survival during cellular stress and carbon source limitation. The diverse presence of this new pathway, toward 2-methylbutanoic acid, across various domains or species (L. lactis, Clostridium spp., and Pseudomonas spp.) may support life under carbohydrate deprivation and stress conditions (13, 18, 31). Leucine catabolism to 3-methylbutanoic and 2-methylbutanoic acids, by Ganesan's pathway, can serve these purposes also in L. sanfranciscensis LSCE1, suggesting that the switch from sugar to amino acid catabolism supports survival and growth also in specific and restricted environments, such as sourdoughs, characterized by acid stress and recurrent carbon starvation (8, 9, 18, 42).
ACKNOWLEDGMENT
Luciana Perillo is acknowledged for precious technical assistance.
Footnotes
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Arendt E. K., Ryan L. A. M., Dal Bello F. 2007. Impact of sourdough on the texture of bread. Food Microbiol. 24:165–174 [DOI] [PubMed] [Google Scholar]
- 2. Barber B., Ortola C., Barber S., Fernandez F. 1992. Storage of packaged white bread. III. Effects of sourdough and addition of acids on bread characteristics. Z. Lebensm. Unters. Forsch. 194:442–449 [Google Scholar]
- 3. Beck H. C., Hansen A. M., Lauritsen F. R. 2004. Catabolism of leucine to branched-chain fatty acids in Staphylococcus xylosus. J. Appl. Microbiol. 96:1185–1193 [DOI] [PubMed] [Google Scholar]
- 4. Bore E., Langsrud S., Langsrud Ø., Rode T. M., Holck A. 2007. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153:2289–2303 [DOI] [PubMed] [Google Scholar]
- 5. Chambellon E., et al. 2009. The d-2-hydroxyacid dehydrogenase incorrectly annotated PanE is the sole reduction system for branched-chain 2-keto acids in Lactococcus lactis. J. Bacteriol. 191:873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corsetti A., Settanni L. 2007. Lactobacilli in sourdough fermentation. Food Res. Int. 40:539–558 [Google Scholar]
- 7. Cotter P. D., Hill C. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Angelis M., Bini L., Pallini V., Cocconcelli P. S., Gobbetti M. 2001. The acid-stress response in Lactobacillus sanfranciscensis CB1. Microbiology 147:1863–1873 [DOI] [PubMed] [Google Scholar]
- 9. De Angelis M., Gobbetti M. 2004. Environmental stress responses in Lactobacillus: a review. Proteomics 4:106–122 [DOI] [PubMed] [Google Scholar]
- 10. De Vuyst L., Vancanneyt M. 2007. Biodiversity and identification of sourdough lactic acid bacteria. Food Microbiol. 24:120–127 [DOI] [PubMed] [Google Scholar]
- 11. De Vuyst L., Vrancken G., Ravyts F., Rimaux T., Weckx S. 2009. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 26:666–675 [DOI] [PubMed] [Google Scholar]
- 12. Di Cagno R., et al. 2002. Proteolysis by sourdough lactic acid bacteria: effects on wheat flour protein fractions and gliadin peptides involved in human cereal intolerance. Appl. Environ. Microbiol. 68:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elsden S. R., Hilton M. G. 1978. Volatile acid production from threonine, valine, leucine and isoleucine by clostridia. Arch. Microbiol. 117:165–172 [DOI] [PubMed] [Google Scholar]
- 14. Erasmus D. J., K. van der Merwe G., van Vuuren H. J. 2003. Genome-wide expression analyses: metabolic adaptation of Saccharomyces cerevisiae to high sugar stress. FEMS Yeast Res. 3:375–399 [DOI] [PubMed] [Google Scholar]
- 15. Fernández M., Kleerebezem M., Kuipers O. P., Siezen R. J., van Kranenburg R. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrara C. T., et al. 2008. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 4:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fleige S., et al. 2006. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol. Lett. 28:1601–1613 [DOI] [PubMed] [Google Scholar]
- 18. Ganesan B., Dobrowolski P., Weimer B. C. 2006. Identification of the leucine-to-2-methylbutyric acid catabolic pathway of Lactococcus lactis. Appl. Environ. Microbiol. 72:4264–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gänzle M. G., Vermeulen N., Vogel R. F. 2007. Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough. Food Microbiol. 24:128–138 [DOI] [PubMed] [Google Scholar]
- 20. Gänzle M. G., Zhang C., Monang B. S., Lee V., Schwab C. 2009. Novel metabolites from cereal-associated lactobacilli—novel functionalities for cereal products? Food Microbiol. 26:712–719 [DOI] [PubMed] [Google Scholar]
- 21. Gianotti A., Serrazanetti D. I., Sado Kamdem S. L., Guerzoni M. E. 2008. Involvement of cell fatty acid composition and lipid metabolism in adhesion mechanism of Listeria monocytogenes. Int. J. Food Microbiol. 123:9–17 [DOI] [PubMed] [Google Scholar]
- 22. Gobbetti M., Corsetti A. 1997. Lactobacillus sanfrancisco, a key sourdough lactic acid bacterium: a review. Food Microbiol. 14:175–187 [Google Scholar]
- 23. Guerzoni M. E., Vernocchi P., Ndagijimana M., Gianotti A., Lanciotti R. 2007. Generation of aroma compounds in sourdough: effects of stress exposure and lactobacilli-yeasts interactions. Food Microbiol. 24:139–148 [DOI] [PubMed] [Google Scholar]
- 24. Hammes W. P., et al. 2005. Microbial ecology of cereal fermentations. Trends Food Sci. Technol. 16:4–11 [Google Scholar]
- 25. Hörmann S., et al. 2006. Comparative proteome approach to characterize the high-pressure stress response of Lactobacillus sanfranciscensis DSM 20451T. Proteomics 6:1878–1885 [DOI] [PubMed] [Google Scholar]
- 26. Hüfner E., Britton R. A., Roos S., Jonsson H., Hertel C. 2008. Global transcriptional response of Lactobacillus reuteri to the sourdough environment. Syst. Appl. Microbiol. 31:323–338 [DOI] [PubMed] [Google Scholar]
- 27. Jänsch A., Korakli M., Vogel R. F., Gänzle M. G. 2007. Glutathione reductase from Lactobacillus sanfranciscensis DSM20451T: contribution to oxygen tolerance and thiol exchange reactions in wheat sourdoughs. Appl. Environ. Microbiol. 73:4469–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kline L., Sugihara T. F. 1971. Microorganism of the San Francisco sourdough bread process. II. Isolation and characterization of undescribed bacterial species responsible for the souring activity. Appl. Microbiol. 21:459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lanciotti R., Patrignani F., Iucci L., Saracino P., Guerzoni M. E. 2007. Potential of high-pressure homogenization in the control and enhancement of proteolytic and fermentative activities of some Lactobacillus species. Food Chem. 102:542–550 [Google Scholar]
- 30. Marles-Wright J., Lewis R. 2007. Stress response of bacteria. Curr. Opin. Struct. Biol. 17:755–760 [DOI] [PubMed] [Google Scholar]
- 31. Massey L. K., Conrad R. S., Sokatch J. R. 1974. Regulation of leucine catabolism in Pseudomonas putida. J. Bacteriol. 118:112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montanari C., Sado-Kamdem S. L., Serrazanetti D. I., Etoa F. X., Guerzoni M. E. 2010. Synthesis of cyclopropane fatty acids in Lactobacillus helveticus and Lactobacillus sanfranciscensis and their cellular fatty acids changes following short term acid and cold stresses. Food Microbiol. 27:493–502 [DOI] [PubMed] [Google Scholar]
- 33. Ndagijimana M. 2001. Ph.D. thesis, CF980202070. University of Bologna, Bologna, Italy [Google Scholar]
- 34. Ndagijimana M., et al. 2006. Two 2(5H)-furanones as possible signalling molecules in Lactobacillus helveticus. Appl. Environ. Microbiol. 72:6053–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olier M., et al. 2004. Screening of glutamate decarboxylase activity and bile salt resistance of human asymptomatic carriage, clinical, food, and environmental isolates of Listeria monocytogenes. Int. J. Food Microbiol. 93:87–99 [DOI] [PubMed] [Google Scholar]
- 36. Patrignani F., et al. 2008. Effects of sub-lethal concentrations of hexanal and 2-(E)-hexenal on membrane fatty acid composition and volatile compounds of Listeria monocytogenes, Staphylococcus aureus, Salmonella enteritidis and Escherichia coli. Int. J. Food Microbiol. 123:1–8 [DOI] [PubMed] [Google Scholar]
- 37. Pfaffl M. W. 2001. A new mathematical method for relative quantification in real time RT-PCR. Nucleic Acids Res. 29:2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Picozzi C., D'Anchise F., Foschino R. 2006. PCR detection of Lactobacillus sanfranciscensis in sourdough and Panettone baked product. Eur. Food Res. Technol. 222:330–335 [Google Scholar]
- 39. Sado-Kamdem S. L., Vannini L., Guerzoni M. E. 2009. Effect of α-linolenic, capric and lauric acid on the fatty acid biosynthesis in Staphylococcus aureus. Int. J. Food Microbiol. 129:288–294 [DOI] [PubMed] [Google Scholar]
- 40. Sanders J. W., et al. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299–310 [DOI] [PubMed] [Google Scholar]
- 41. Serrazanetti D. I., Guerzoni M. E., Corsetti A., Vogel R. F. 2009. Metabolic impact and potential exploitation of the stress reactions in lactobacilli. Food Microbiol. 26:700–711 [DOI] [PubMed] [Google Scholar]
- 42. Serrazanetti D. I. 2009. Effects of acidic and osmotic stresses on flavour compounds and gene expression in Lactobacillus sanfranciscensis. Ph.D. thesis. University of Teramo, Teramo, Italy [Google Scholar]
- 43. Shepard B. D., Gilmore M. S. 1999. Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed PCR. Appl. Environ. Microbiol. 65:1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Siragusa S., et al. 2007. Synthesis of γ-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl. Environ. Microbiol. 73:7283–7290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smit B., Engels W., Wouters J., Smit G. 2004. Diversity of L-leucine catabolism in various microorganisms involved in dairy fermentations, and identification of the rate-controlling step in the formation of the potent flavour component 3-methylbutanal. Appl. Microbiol. Biotechnol. 64:396–402 [DOI] [PubMed] [Google Scholar]
- 46. Smit B. A., et al. 2005. Identification, cloning, and characterization of a Lactococcus lactis branched-chain α-keto acid decarboxylase involved in flavor formation. Appl. Environ. Microbiol. 71:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van de Guchte M., et al. 2002. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82:187–216 [PubMed] [Google Scholar]
- 48. Vannini L., et al. 2007. New signaling molecules in some gram-positive and gram-negative bacteria. Int. J. Food Microbiol. 120:25–33 [DOI] [PubMed] [Google Scholar]
- 49. Velasco S. E., et al. 2007. Influence of the carbohydrate source on β-glucan production and enzyme activities involved in sugar metabolism in Pediococcus parvulus 2.6. Int. J. Food Microbiol. 115:325–334 [DOI] [PubMed] [Google Scholar]
- 50. Vermeulen N., Gänzle M. G., Vogel R. F. 2007. Glutamine deamidation by cereal-associated lactic acid bacteria. J. Appl. Microbiol. 103:1197–1205 [DOI] [PubMed] [Google Scholar]
- 51. Vernocchi P., et al. 2008. Influence of starch addition and dough microstructure on fermentation aroma production by yeasts and lactobacilli. Food Chem. 108:1217–1225 [Google Scholar]
- 52. Vogel R. F., et al. 1999. Non-dairy lactic fermentations: the cereal world. Antonie Van Leeuwenhoek 76:403–411 [PubMed] [Google Scholar]
- 53. Ward D. E., et al. 2000. Branched-chain α-keto acid catabolism via the gene products of the bkd operon in Enterococcus faecalis: a new, secreted metabolite serving as a temporary redox sink. J. Bacteriol. 182:3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Welch D. F. 1991. Applications of cellular fatty acid analysis. Clin. Microbiol. 4:422–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Williams A. G., Withers S. E., Brechany E. Y., Banks J. M. 2006. Glutamate dehydrogenase activity in lactobacilli and the use of glutamate dehydrogenase-producing adjunct Lactobacillus spp. cultures in the manufacture of cheddar cheese. J. Appl. Microbiol. 101:1062–1075 [DOI] [PubMed] [Google Scholar]
- 56. Yvon M., Thirouin S., Rijnen L., Fromentier D., Gripon J. C. 1997. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl. Environ. Microbiol. 63:414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]