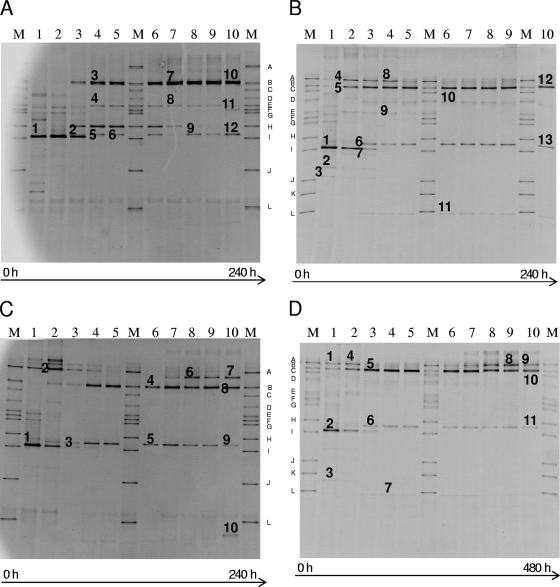
Fig. 3.
Culture-independent 16S rRNA-PCR-DGGE analysis of fermentation samples taken at each refreshment step of a sourdough fermentation backslopped 10 times. Panels A to D correspond to fermentations A to D in Table 1. The marker ladder M consisted of Lactobacillus plantarum LMG 6907T (band A), Lactobacillus fermentum LMG 6902T (band B), Leuconostoc mesenteroides LMG 6893T (band C), Lactobacillus brevis LMG 6906T (band D), Lactobacillus curvatus LMG 69198T (band E), Pediococcus pentosaceus LMG 10478 (band F), Enterococcus faecalis LMG 7937T (band G), Lactococcus lactis LMG 19152T (band H), Lactobacillus sanfranciscensis 16002T (band I), Bifidobacterium longum BB536 (band J), Bifidobacterium adolescentis LMG 10502 (band K), and Bifidobacterium breve LMG 11084 (band L). Bands indicated by numbers have been excised and subjected to 16S rRNA gene sequence analysis. The closest relatives of the fragments sequenced are listed in Table 1.