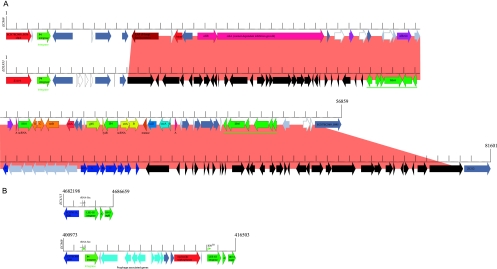
Fig. 3.
Prophage polymorphisms. (A) Strains EC869 and EDL933 share parts of P4 prophages targeting the clpA locus, which in the case of EC869 introduces loci that are intimately associated with host prevalence and pathogenic potential. The phage architecture in EC869 is distinguished by a 56,859-kb insertion, featuring a Tn10-type transposon. This lateral acquired region carries 57 genes (ECH7EC869_5838 to _5894), mediating contact-dependent inhibition of growth (cdiAB), adhesion, and resistance to streptomycin (strAB), tetracyclines (tetDBAR), sulfonamides (sul) (dark blue), and cobalt-zinc-cadmium (czcAB) (light blue). To our knowledge this is the first report of czc resistance in E. coli. Corresponding loci are colored accordingly, with hypothetical proteins (light gray), conserved hypothetical proteins (dark gray), pseudogenes (white), mobile elements (dark green), and adhesins (purple). In the phage of strain EDL933, dark blue marks the tetracycline resistance operon. (B) Strain EC869 shows polymorphisms within the borders of the LEE pathogenicity island caused by the integration of an 11,098-bp P4 prophage remnant targeting the selC locus (ECH74115_5031). This novel phage remnant was also identified in the draft contigs of the FRIK2000 and FRIK966 genomes, supporting their SNP-derived phylogenetic placement. The integration side is a likely target for lateral acquisition, as evidenced by its deviating GC content linked to the AT-rich LEE-borne phage integrase and neighboring IS911 element. Integration results in a 30-bp imperfect direct repeat (IDR; arrows) and introduces 12 genes (ECH7EC869_1671 to _1682). Corresponding loci are colored accordingly, with hypothetical proteins (light gray) and conserved hypothetical proteins (dark gray).